For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDRICRLALSHVVLPLPKPVSDAKVLTGRQKPLTETVLLFVEVSTSDGLQGMGFSYSKRAGGPAQYTHLH EIAEVALGQDPSDIDRIYQSLLWAGASVGRSGVATQAIAALDVALWDLKARRAGLPLAKLIGAQRDSCRV YNTSGGFLQASLDEVKDAATASLESGIGGVKIKVGQPDWAEDLKRVAAMRAHLGDTPLMVDANQQWDRAR ARRMCRELEQFDLVWIEEPLDAWDAVGHADLSHTFDTPIATGEMLTSVPEHMALIDAGYRGIVQPDAPRI GGITPFLRFATLASHAGLALAPHYAMEIHLHLAATYPTEPWVEHFEWLNPLFNERIEIRDGRMWLPDRPG LGFTLSDQMRALTREQAEFQA*
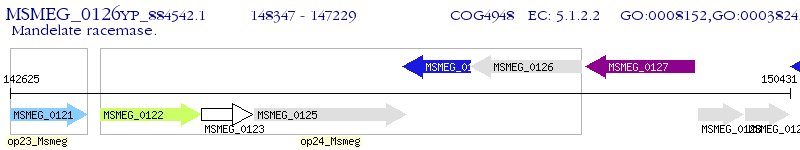
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0126 | - | - | 100% (372) | mandelate racemase/muconate lactonizing enzyme |
| M. smegmatis MC2 155 | MSMEG_6177 | - | 2e-26 | 28.61% (346) | galactonate dehydratase |
| M. smegmatis MC2 155 | MSMEG_2909 | - | 1e-25 | 26.23% (366) | starvation-sensing protein RspA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1357 | - | 5e-15 | 26.82% (261) | mandelate racemase/muconate lactonizing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0864 | - | 9e-19 | 29.47% (285) | mandelate racemase |
| M. avium 104 | MAV_3096 | - | 7e-33 | 31.10% (344) | mandelate racemase/muconate lactonizing enzyme |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1017 | - | 4e-13 | 24.25% (400) | glucarate dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0126|M.smegmatis_MC2_155 MTDRICRLALSHVVLPLPKPVSDAKVLTGRQKPLTETVLLFVEVSTSDGL
MAV_3096|M.avium_104 MTPDPAIDEVTARVYQIPTDAPEADGTLS----WSSTTLVLAEVAAG-GR
Mflv_1357|M.gilvum_PYR-GCK --MKITAVDAIPFAIPYTKPLKFASGEVH------TAEHVLVRVHTDEGV
Mvan_1017|M.vanbaalenii_PYR-1 MRTPVITDVTVVPIAGHDSMLLNLSGAHG-----PFFTRNLVIVEDSEGN
MMAR_0864|M.marinum_M MRIVAILERPVRLLGGPANAVVNFSQHTVSLVAVVSDVIRHGKPLTGVAF
. . . .
MSMEG_0126|M.smegmatis_MC2_155 QGMG-FSYSKRAGGPAQYTHLHEIAEVALGQDPSDIDRIYQSLLWAGA--
MAV_3096|M.avium_104 RGIG-YTY---ADGACAALITGALAGSVTGHSALDVPARWQAMVRAMR--
Mflv_1357|M.gilvum_PYR-GCK VGVA-EAPP--RPFTYGETQDGILAVIRQIFAPQIVGLTLTEREVVNA--
Mvan_1017|M.vanbaalenii_PYR-1 TGVG-EVPGGEPIRRTLQDARGIVTGRSIGDYHAVLNEMRRTFADRDAGG
MMAR_0864|M.marinum_M NSIGRFAQSGILRDRMIPRVLAAPEDALLDQRGHLDPAAVLRHAVCDE--
.:.
MSMEG_0126|M.smegmatis_MC2_155 ---SVGRSGVATQAIAALDVALWDLKARRAGLPLAKLIGAQRDS-----C
MAV_3096|M.avium_104 ---NNGRPGLVSCAISAVDTALWDLKGKLLGLPVCRLLGMAHDS-----V
Mflv_1357|M.gilvum_PYR-GCK ---RLARTVGNPTAKAAVDMAIWDALGRSLDLSVSALLGGYGDH-----M
Mvan_1017|M.vanbaalenii_PYR-1 RGAQTFDLRVTVHAVTAIESALMDLLGQHLEVPVAALLGDGQQR-----S
MMAR_0864|M.marinum_M ---KPGGHGDRAGAAAAVELACWDLAAKLNDEPAYATIARHFGRDPAPLV
* :*:: * * .: . :.
MSMEG_0126|M.smegmatis_MC2_155 RVYNTSGGFLQASLD--------------------------------EVK
MAV_3096|M.avium_104 PIYGSGGFTSYDDNA--------------------------------TRA
Mflv_1357|M.gilvum_PYR-GCK RVSHMLG--FDTPDT--------------------------------MVA
Mvan_1017|M.vanbaalenii_PYR-1 RVQALGYLFFVGDRTRTDLAYRSPSDEEPGADDWFTVRHEEAMTPDAVVR
MMAR_0864|M.marinum_M SVYAAGGYYYPGGGLN-------------------------------ALR
:
MSMEG_0126|M.smegmatis_MC2_155 DAATASLESGIGGVKIKVGQ---PDWAEDLKRVAAMRAHLG-DTPLMVDA
MAV_3096|M.avium_104 QLERWVHEWRIPRVKIKIGESWGSHERRDLDRIALARTVIGPDTELYVDA
Mflv_1357|M.gilvum_PYR-GCK EAERIRDEYGISTFKVKVGR---RPVTLDTAVVRALRERFGDTVELYVDG
Mvan_1017|M.vanbaalenii_PYR-1 LAEAARARYGFADFKLKGGV---LPAADEAKAVIALAERFP-DSRITLDP
MMAR_0864|M.marinum_M DEMRRYLDLGYDAVKMKIGG---VAQRDDLARIEAVIDVVG-ADRVAVDA
.*:* * : : . : :*
MSMEG_0126|M.smegmatis_MC2_155 NQQWDRARARRMCRELEQFDLVWIEEPLDAWDAVGHADLSHTFDTPIATG
MAV_3096|M.avium_104 NGGYRRKQAIRMAHAMAGHDVTWFEEPVSSDDLAGLREVRDQVSADVTAG
Mflv_1357|M.gilvum_PYR-GCK NRGWTASESLRALKDMADLDLLFAEELCPADDVLGRRWLVEHTEIPFIAD
Mvan_1017|M.vanbaalenii_PYR-1 NGGWLLADAIRTCRELKDV-LAYAEDPVGPEDGFSGREVMAEFKRATGLP
MMAR_0864|M.marinum_M NGRFDRAAATRWAATLAPYGLRWYEEPGDPLDFELNRAVTGCYAGTVATG
* : : * : : : *: . * :
MSMEG_0126|M.smegmatis_MC2_155 EMLTSVPEHMALIDAG-YRG---IVQPDAPRIGGITPFLRFATLASHAGL
MAV_3096|M.avium_104 EYGYDLAYFHRMLAAD-AVD---CLQIDVTRCGGITDWLRAAAVAAAHNV
Mflv_1357|M.gilvum_PYR-GCK ESVPTPADVTREVLGG-SAT---AISIKTARTG-FTGSRRVHHLAEGLGL
Mvan_1017|M.vanbaalenii_PYR-1 TATNMIATDWREMGHA-IRSGAVDIPLADPHFWTMTGSVRVAQLCDAWGL
MMAR_0864|M.marinum_M ENLFSVPDVTNLVRYGGMRAGRDIFQMDAGLSYGLGEYVRMLELIEAHGF
. : . : * : ..
MSMEG_0126|M.smegmatis_MC2_155 ALAP--------HYAMEIHLHLAATYPTEPWVEHFEWLN---PLFNERIE
MAV_3096|M.avium_104 DVSG--------HCAPNLHAHVATAVANLRHLEYFHDHHRIERMFFDGAL
Mflv_1357|M.gilvum_PYR-GCK EVVMGNQIDGQLGTACAVAFGAAFELTSRRAGELSNFLDMSDDLLTHPLR
Mvan_1017|M.vanbaalenii_PYR-1 TWGSHSNNHFDVSLAMFTHVAAAAPGDITAIDTHWIWQD-GQAITTNPYP
MMAR_0864|M.marinum_M DRRCAYP-----HGGHMINLHIAAGLG-LGGCESYPGVFQPFGGYCDGCV
. * .
MSMEG_0126|M.smegmatis_MC2_155 IRDGRMWLPDRPGLGFTLSDQMRALTREQAEFQA----------------
MAV_3096|M.avium_104 SPQGGVLRPDQQRPGFGLEFKQRDADRYRMR-------------------
Mflv_1357|M.gilvum_PYR-GCK IRDGRLHVP--PGPGLGIEIDPDKLDHYRTDH------------------
Mvan_1017|M.vanbaalenii_PYR-1 IVDGYLSVPDAPGLGVTLDEPAVQAAHALYQREGLGGRDDAVAMQYLIPG
MMAR_0864|M.marinum_M LRDGQIVPTDAPGFGLERKPGLGELISELTR-------------------
:* : . *. .
MSMEG_0126|M.smegmatis_MC2_155 -------------
MAV_3096|M.avium_104 -------------
Mflv_1357|M.gilvum_PYR-GCK -------------
Mvan_1017|M.vanbaalenii_PYR-1 WTFDGKRPALDRG
MMAR_0864|M.marinum_M -------------