For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPGQPSTDPVVPRARVANLANVLTGIRMALVPVFLVFLFTDGGHEFFWRMAAFTVFAVAVITDRFDGALA RSYGMVTEFGALADPIADKALIGAALIGLSMLGDLPWWVTAVVLAREIGITLMRLAVLRHGVIPASRGGK LKTTVQAVAIGLFVMPLYAWSPVWTTVAWAIMWAAVVLTVLTGADYVVSAIRDSRGRPADN
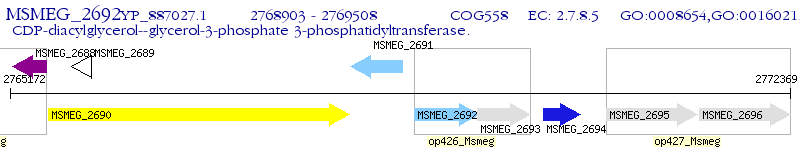
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2692 | pgsA | - | 100% (201) | CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase |
| M. smegmatis MC2 155 | MSMEG_3652 | - | 2e-14 | 30.60% (183) | CDP-diacylglycerol-glycerol-3-phosphate |
| M. smegmatis MC2 155 | MSMEG_2933 | - | 1e-04 | 25.89% (197) | phosphatidylinositol synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2767c | pgsA3 | 2e-65 | 63.27% (196) | CDP-diacylglycerol--glycerol-3-phosphate |
| M. gilvum PYR-GCK | Mflv_3989 | - | 1e-82 | 77.95% (195) | CDP-diacylglycerol--glycerol-3-phosphate |
| M. tuberculosis H37Rv | Rv2746c | pgsA3 | 2e-65 | 63.27% (196) | CDP-diacylglycerol--glycerol-3-phosphate |
| M. leprae Br4923 | MLBr_00979 | - | 2e-65 | 65.78% (187) | CDP-diacylglycerol--glycerol-3-phosphate |
| M. abscessus ATCC 19977 | MAB_3071c | - | 2e-62 | 60.71% (196) | PGP synthase PgsA3 (phosphatidylglycerophosphate synthase) |
| M. marinum M | MMAR_1969 | pgsA3 | 1e-65 | 68.68% (182) | PGP synthase PgsA3 |
| M. avium 104 | MAV_3637 | pgsA | 4e-62 | 62.37% (186) | CDP-diacylglycerol--glycerol-3-phosphate |
| M. thermoresistible (build 8) | TH_0577 | pgsA3 | 6e-83 | 80.98% (184) | PROBABLE PGP SYNTHASE PGSA3 |
| M. ulcerans Agy99 | MUL_3388 | pgsA3 | 4e-68 | 66.67% (195) | PGP synthase PgsA3 |
| M. vanbaalenii PYR-1 | Mvan_2395 | - | 2e-83 | 78.39% (199) | CDP-diacylglycerol--glycerol-3-phosphate |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2692|M.smegmatis_MC2_155 ------------------------MPGQPSTDPVVPRARVANLANVLTGI
TH_0577|M.thermoresistible__bu ---------------------------------------VANLANALTGV
Mflv_3989|M.gilvum_PYR-GCK ----------------MCNQYSVAVPGQPHTDPVVPRARVANIANVLTGI
Mvan_2395|M.vanbaalenii_PYR-1 ----------------------MAVPGQPHTDPVVPRARVANLANLLTGI
MLBr_00979|M.leprae_Br4923 ---------------------------------MVDCVRIANLANILTGL
MAV_3637|M.avium_104 ---------------------------------MVGRANIANLANTLTLL
Mb2767c|M.bovis_AF2122/97 --------MSRS------TRYSVAVSAQPETGQIAGRARIANLANILTLL
Rv2746c|M.tuberculosis_H37Rv --------MSRS------TRYSVAVSAQPETGQIAGRARIANLANILTLL
MMAR_1969|M.marinum_M ---------------------------------MPGTAGIANLANVLTGL
MUL_3388|M.ulcerans_Agy99 MTFGHEKTVSTSAAGRLVNRYSVAVSGQPQTGQVPGTVGIANLANVLTGL
MAB_3071c|M.abscessus_ATCC_199 --------------------MPMPPETQPDSAASAAPVPVLNIANILTVL
: *:** ** :
MSMEG_2692|M.smegmatis_MC2_155 RMALVPVFLVFLFTDGGHEFFWRMAAFTVFAVAVITDRFDGALARSYGMV
TH_0577|M.thermoresistible__bu RLLLVPVFLILLFVGDGHETSWRIGAFAVFAVAVITDRFDGALARSYGMV
Mflv_3989|M.gilvum_PYR-GCK RMALVPVFLVALFVGDGHETFWRLTAFTIFLVAVITDRLDGALARSYGMV
Mvan_2395|M.vanbaalenii_PYR-1 RMALVPVFLFALFAGDGQETSWRITAFVVFAVAVITDRFDGALARSYGMV
MLBr_00979|M.leprae_Br4923 RLMLVPVFLVALFAGNGHEIVGRIAAFVIFTVACITDRLDGLLARRYGMA
MAV_3637|M.avium_104 RLVLVPVFLLALFAGDGHETGFRILAFLIFAFACITDRLDGLLARNYGMA
Mb2767c|M.bovis_AF2122/97 RLVMVPVFLLALFYGGGHHSAARVVAWAIFATACITDRFDGLLARNYGMA
Rv2746c|M.tuberculosis_H37Rv RLVMVPVFLLALFYGGGHHSAARVVAWAIFATACITDRFDGLLARNYGMA
MMAR_1969|M.marinum_M RFVLVPIFLYLLFYDGGHQTTFRIAAWAVFAVASITDRFDGLLARNYGMA
MUL_3388|M.ulcerans_Agy99 RFVLVPIFLYLLFYDGGHQTTFRIAAWAVFAVASITDRFDGLLARNYGMA
MAB_3071c|M.abscessus_ATCC_199 RIVLVPVFVVVLFTEGGHQSSWRIAAFGVFALAIATDRYDGILARRYGLI
*: :**:*: ** .*:. *: *: :* * *** ** *** **:
MSMEG_2692|M.smegmatis_MC2_155 TEFGALADPIADKALIGAALIGLSMLGDLPWWVTAVVLAREIGITLMRLA
TH_0577|M.thermoresistible__bu TEFGTLADPIADKALIGAALIGLSLLGDLPWWVTAVILVREIGITALRFA
Mflv_3989|M.gilvum_PYR-GCK TEFGTLADPIADKMLIGSALVGLSILGDLPWWVTVVILVREVAVTVLRLV
Mvan_2395|M.vanbaalenii_PYR-1 TEFGTLADPIADKALIGSALVGLSVLGDLPWWVTVVILVRELGVTLLRLV
MLBr_00979|M.leprae_Br4923 TEFGAFVDPIADKTLIGAALIGLSMLGDLPWWVTVLIMTRELGVTLLRLA
MAV_3637|M.avium_104 TEFGAFVDPIADKTLVGSALIGLSMLGELPWWITVLILAREFGVTVLRLA
Mb2767c|M.bovis_AF2122/97 TEFGAFVDPIADKTLIGSALIGLSMLGDLPWWVTVLILTRELGVTVLRLA
Rv2746c|M.tuberculosis_H37Rv TEFGAFVDPIADKTLIGSALIGLSMLGDLPWWVTVLILTRELGVTVLRLA
MMAR_1969|M.marinum_M TEFGAFVDPIADKALIGAALIGLSMLNDLPWWVTVLILSRELGVTMLRLA
MUL_3388|M.ulcerans_Agy99 TEFGAFVDPIADKALIGAALIGLSMLNDLPWWVTVLILSRELGVTMLRLA
MAB_3071c|M.abscessus_ATCC_199 TPFGQLADPIADKALICAALVGLSLLGDLSWWVTAVILVREIGITVLRFS
* ** :.****** *: :**:***:*.:*.**:*.::: **..:* :*:
MSMEG_2692|M.smegmatis_MC2_155 VLRHGVIPASRGGKLKTTVQAVAIGLFVMPLYAWSPVWTTVAWAIMWAAV
TH_0577|M.thermoresistible__bu VLRHGVIPASRGGKLKTLAQAVAIGLFVLPLANWAPVWSTVAWGIMAVAV
Mflv_3989|M.gilvum_PYR-GCK VLRRGVIPASRGGKIKTLVQAVAIGLFVMPLS---GAWLTGAWVIMAAAG
Mvan_2395|M.vanbaalenii_PYR-1 VLRRGVIPASRGGKLKTLVQAVAIGLFVLPLS---DGWLTGAWVIMLAAV
MLBr_00979|M.leprae_Br4923 VIHRGVIPASWGGKLKTVVQAVAIGLFLLPLTQLSVPFHVGAAVVMAVAV
MAV_3637|M.avium_104 VIRRGVIPASWGGKLKTVVQVFAIGLFVLPWAGTPGPFRVGAAVVMGAAI
Mb2767c|M.bovis_AF2122/97 VIRRGVIPASWGGKLKTFVQAVAIGLFVLPLSG---PLHVAAVVVMAAAI
Rv2746c|M.tuberculosis_H37Rv VIRRGVIPASWGGKLKTFVQAVAIGLFVLPLSG---PLHVAAVVVMAAAI
MMAR_1969|M.marinum_M VIRRGVIPASWGGKLKTVVQAVAIGLFVLPLSG---PLHMAAVVVMAAAI
MUL_3388|M.ulcerans_Agy99 VIRRGVIPASWGGKLKTVVQAVAIGLFVLPLSG---PLHMAAVVVMAAAI
MAB_3071c|M.abscessus_ATCC_199 VLRHGVIPANRGGKLKTLVQSLAIAGYLLPLTG---HWHTAALILMGLAV
*:::*****. ***:** .* .**. :::* * :* *
MSMEG_2692|M.smegmatis_MC2_155 VLTVLTGADYVVSAIRDSRGRPADN-
TH_0577|M.thermoresistible__bu VLTVFTGVDYVVSAVRDSRGRSAGH-
Mflv_3989|M.gilvum_PYR-GCK VLTVLTGVDYFVSAVRDWRRQSPGR-
Mvan_2395|M.vanbaalenii_PYR-1 ALTVTTGVDYFVSAVRDWRGRSAGQ-
MLBr_00979|M.leprae_Br4923 VLTVVTGVDYVAGTIRAVRGVHQTAS
MAV_3637|M.avium_104 VLTVITGIDYVVSTVREVR---RTAA
Mb2767c|M.bovis_AF2122/97 LLTVITGVDYVARALRDIGGIRQTAS
Rv2746c|M.tuberculosis_H37Rv LLTVITGVDYVARALRDIGGIRQTAS
MMAR_1969|M.marinum_M VLTVVTGIDYVASTVREIRAGKRAS-
MUL_3388|M.ulcerans_Agy99 VLTVVTGIDYVASTVREIRAGKRAS-
MAB_3071c|M.abscessus_ATCC_199 ALTVITGIDYLVQALRPRPKTP----
*** ** **.. ::*