For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKPPVTEAVTGEFSYDPFDPAVMADPTPYYRVLREKYPLYHIEKWDTWALSRFSDIWEVLEVNDGTFVAS EGTLPAATVLAEHNDGAVPDPPWHPMPFHANFDTPIYDSVRRCTSPQFRPRSVTKLADRIRTLANERLDE LLPRGQFDLTQDYGGIVAASVVCELVGLPVDLAADVLATVNAGSLAEPGSGVEVANARPGYLEYLTPVVQ RRRAGEGRELPIVDNLLQYRLPDGSAFSDLEAAVQMLGVFIGGTETVPKIVAHGLWELTRHPEQLAAVRT DLDANVPIAREEMIRFCAPAQWFARTVRKPFTLHGTTLQPGQRVITLLASAGRDEREYPDPDSFIWDRRI ERLLAFGRGQHFCLGVHLARLEITIMVTEWLKRVGEFRVDERAARRPPSSFQWGWNNVPVEV
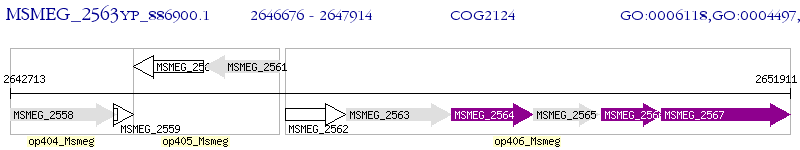
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2563 | - | - | 100% (412) | cytochrome P450 superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5861 | - | 1e-46 | 31.42% (401) | cytochrome P450 109 |
| M. smegmatis MC2 155 | MSMEG_6622 | - | 9e-45 | 32.27% (409) | cytochrome P450 monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0789c | cyp123 | 1e-44 | 31.28% (406) | cytochrome P450 123 |
| M. gilvum PYR-GCK | Mflv_4107 | - | 0.0 | 81.91% (398) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv0766c | cyp123 | 1e-44 | 31.28% (406) | cytochrome P450 123 |
| M. leprae Br4923 | MLBr_02088 | - | 7e-34 | 28.36% (409) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1216c | - | 1e-44 | 30.36% (415) | cytochrome P450 |
| M. marinum M | MMAR_0399 | cyp191A3 | 1e-173 | 70.75% (400) | cytochrome P450 191A3 Cyp191A3 |
| M. avium 104 | MAV_5123 | - | 1e-171 | 69.90% (402) | cytochrome P450 superfamily protein |
| M. thermoresistible (build 8) | TH_3299 | - | 1e-173 | 72.32% (401) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_1049 | cyp191A3 | 1e-169 | 69.50% (400) | cytochrome P450 191A3 Cyp191A3 |
| M. vanbaalenii PYR-1 | Mvan_2239 | - | 0.0 | 81.75% (400) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4107|M.gilvum_PYR-GCK ------------------------------------MRYDPFDPAVMADP
Mvan_2239|M.vanbaalenii_PYR-1 ---------------------MSAAQQPDCRPGRRPFTYDPFDPDVMADP
MSMEG_2563|M.smegmatis_MC2_155 ------------------------MKPPVTEAVTGEFSYDPFDPAVMADP
TH_3299|M.thermoresistible__bu ------------------------MTVSHETFSHETFSYDPFDPDVMADP
MMAR_0399|M.marinum_M ----------------------------------MNFSYDPFGARVMADP
MUL_1049|M.ulcerans_Agy99 ----------------------------------MNFSYDPFGARVMADP
MAV_5123|M.avium_104 ----------------------------------MDFAYDPFDAEVMANP
Mb0789c|M.bovis_AF2122/97 --------------------------MTVRVGDPE-LVLDPYDYDFHEDP
Rv0766c|M.tuberculosis_H37Rv --------------------------MTVRVGDPE-LVLDPYDYDFHEDP
MAB_1216c|M.abscessus_ATCC_199 --------------------------MTVAETSAVPLVFDPYDYDFHEDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
: . :*
Mflv_4107|M.gilvum_PYR-GCK LPYYRVLRDEHPVHYLETWDTYALSRFDDIWNVLEINDGTFVASEGTLPA
Mvan_2239|M.vanbaalenii_PYR-1 LPYYRVLRDEHPVYYLQKWDTYALSRFDDIWNVLEINDGTFVASEGTLPS
MSMEG_2563|M.smegmatis_MC2_155 TPYYRVLREKYPLYHIEKWDTWALSRFSDIWEVLEVNDGTFVASEGTLPA
TH_3299|M.thermoresistible__bu LPYYRVLRDRHPVYHVAKWDTYALSRFEDIWQVLEVNDGTFVASEGTLPP
MMAR_0399|M.marinum_M LPYYRVLRDQHPVYYLPQWDTFAISRFDDIWRVLEVNDGTFVASEGTLPA
MUL_1049|M.ulcerans_Agy99 LPYYRVLRDQHPVYYLPQWDTFAISRFDDIWRVLEVNDGTFVASEGTLPA
MAV_5123|M.avium_104 LPYYRILRDHHPVYYMPQWDTFALSRFDDIWRVLEVNNGTFVASEGTLPP
Mb0789c|M.bovis_AF2122/97 YPYYRRLRDEAPLYRNEERNFWAVSRHHDVLQGFRDSTALSNAYGVSLDP
Rv0766c|M.tuberculosis_H37Rv YPYYRRLRDEAPLYRNEERNFWAVSRHHDVLQGFRDSTALSNAYGVSLDP
MAB_1216c|M.abscessus_ATCC_199 YPYYRRLRDEAPLYRNDDLKFWALSRHHDVLQGFRNSEALSNANGVSMDK
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPG-MPLTVFSSFSDCDEALRHPLSASDRLKATLAQ
* ** * : *: ..* . * . :. . ::
Mflv_4107|M.gilvum_PYR-GCK AAVLARHNDGPVADPPLHPMPFHANFDSPIYEDVRRCTATPFRPKNVAKL
Mvan_2239|M.vanbaalenii_PYR-1 AAVLAGHNDGPVADPPLHPMPFHANFDAPIYDEVRRCTAAPFRPRNVAKL
MSMEG_2563|M.smegmatis_MC2_155 ATVLAEHNDGAVPDPPWHPMPFHANFDTPIYDSVRRCTSPQFRPRSVTKL
TH_3299|M.thermoresistible__bu ATTLARHNSGPVPDPPLQPLPFHAVFDAPLYNSVRRTHSAPFRPRSVAGM
MMAR_0399|M.marinum_M ASVLARHNSGPVDDPPWHPLPFHAVFDAPLYGEIRRAHSPPLRPKSVAGL
MUL_1049|M.ulcerans_Agy99 ALVLARHNSGPVDDPPWHPLPFRAVFDAPLCGEIRRAHSPPLRPKSVAGL
MAV_5123|M.avium_104 ASVLAQHNDGPVDDPPLHPLPFHAMFDADLYGEIRRTHSRPFRPRAVTDL
Mb0789c|M.bovis_AF2122/97 SSRTS----------EAYRVMSMLAMDDPAHLRMRTLVSKGFTPRRIREL
Rv0766c|M.tuberculosis_H37Rv SSRTS----------EAYRVMSMLAMDDPAHLRMRTLVSKGFTPRRIREL
MAB_1216c|M.abscessus_ATCC_199 ASFGP----------HAKLVMSFLAMDDPEHLRLRALVSKGFTPRRIREL
MLBr_02088|M.leprae_Br4923 QAIAAGA------EPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQAL
. :* :* : : *: : :
Mflv_4107|M.gilvum_PYR-GCK RDRIRVLANERLDELLPLGR---FDLTQDYGGIVAASVVCELVGLPAEMA
Mvan_2239|M.vanbaalenii_PYR-1 ADRIRVLANERLDELLPRGR---FDLTQEYGGIVAASVVCELVGLPADLA
MSMEG_2563|M.smegmatis_MC2_155 ADRIRTLANERLDELLPRGQ---FDLTQDYGGIVAASVVCELVGLPVDLA
TH_3299|M.thermoresistible__bu RDRIRELATERLDALLPRGT---FDLTQEYAGVVAAAMVCDLLGVPTKLA
MMAR_0399|M.marinum_M EARIRELANERLDRLLPQGS---FDLTQDYGGVVVASIVCELLGMPASLA
MUL_1049|M.ulcerans_Agy99 EARIRELANERLNRLLPQGS---FDLTQDYGGVVVASIVCELLGMPASLA
MAV_5123|M.avium_104 EGRIRTLANERLDELLARGS---FDLTQEYGGVVVATIVCELLGIPTDLA
Mb0789c|M.bovis_AF2122/97 EPQVLELARIHLDSALQTES---FDFVAEFAGKLPMDVISELIGVPDTDR
Rv0766c|M.tuberculosis_H37Rv EPQVLELARIHLDSALQTES---FDFVAEFAGKLPMDVISELIGVPDTDR
MAB_1216c|M.abscessus_ATCC_199 EGQVVALARTHLDRALANASDRSFDFIAEYAGKLPMDVISELMGVPESDR
MLBr_02088|M.leprae_Br4923 EGDIAALVDSLLDKGAAAGQ---FDVIADLAFPLAVAVICRLLGVPYEDA
: *. *: **. : . : ::. *:*:*
Mflv_4107|M.gilvum_PYR-GCK ADVLATVNAGSLAQPGSGVEV----------ANARPGYLEYLTPVVERTR
Mvan_2239|M.vanbaalenii_PYR-1 PDVLATVNAGSLAQPGSGVEV----------ANARPGYLEYLTPVVARRR
MSMEG_2563|M.smegmatis_MC2_155 ADVLATVNAGSLAEPGSGVEV----------ANARPGYLEYLTPVVQRRR
TH_3299|M.thermoresistible__bu PDVLATVNAGSLPQQDKGVDT----------AAARPGYLEYLVPVVQRRR
MMAR_0399|M.marinum_M PEVLAAVNAGSLTEPGVGVDT----------AETRPNYLEFLVPVVRQRR
MUL_1049|M.ulcerans_Agy99 PEVLAAVNAGSLTEPGVGVDT----------AETRPNYLEFLVPVVRQRR
MAV_5123|M.avium_104 PQVLAAVNAGSLAEPGVGVDT----------GQARPNYFEFLLPAVQRRR
Mb0789c|M.bovis_AF2122/97 ARIRALADAVLHREDGVADVP-------PPAMAASIELMRYYADLIAEFR
Rv0766c|M.tuberculosis_H37Rv ARIRALADAVLHREDGVADVP-------PPAMAASIELMRYYADLIAEFR
MAB_1216c|M.abscessus_ATCC_199 ARIRELADGVMHREDGLADVP-------PEAIQASFDLMTYYIEMVRERR
MLBr_02088|M.leprae_Br4923 PEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRR
. . : : : *
Mflv_4107|M.gilvum_PYR-GCK AGEPG-DLPIVDNLLAYRLPDGSALSDMEAAVQMLGVFIGGTETVPKIVA
Mvan_2239|M.vanbaalenii_PYR-1 AGQGS-ELPIVDNLLNYRLPDGSALSDTEAAVQMLGVFIGGTETVPKIVA
MSMEG_2563|M.smegmatis_MC2_155 AGEGR-ELPIVDNLLQYRLPDGSAFSDLEAAVQMLGVFIGGTETVPKIVA
TH_3299|M.thermoresistible__bu AEDADGSLPIVDGLIRYRLPDGRALDDVEVATQMLCVFIGGTETVPKIVA
MMAR_0399|M.marinum_M ADPTG-ELPVVDGLLRYRLPDGSALDDTEVATQMLCIFIGGTETVPKIVA
MUL_1049|M.ulcerans_Agy99 ADPTG-ELPVVDGLLRYRLPDGSALDDTEVATQMLCIFIGGTETVPKIVA
MAV_5123|M.avium_104 ADPSGPPLEVVDGLLGYQLPDGSALDDLEVATQMLCIFIGGTETVPKIVA
Mb0789c|M.bovis_AF2122/97 RRPAN---NLTSALLAAEL-DGDRLSDQEIMAFLFLMVIAGNETTTKLLA
Rv0766c|M.tuberculosis_H37Rv RRPAN---NLTSALLAAEL-DGDRLSDQEIMAFLFLMVIAGNETTTKLLA
MAB_1216c|M.abscessus_ATCC_199 RRPTE---DLTSALLQAEI-DGDRLTDEEVLAFLFLMVIAGNETTTKLLA
MLBr_02088|M.leprae_Br4923 GTPGE---DLISRLIELDE-SGDQLTEEEIIATCGLLLVAGHETTVNLIA
: . *: .* : : * . :.:.* **. :::*
Mflv_4107|M.gilvum_PYR-GCK HGLWELSSRPDQLADVVSDLDANVPVAREEMIRYCAPAQWFARTLRRPFT
Mvan_2239|M.vanbaalenii_PYR-1 HGLWELSSRPDQLAEVRSDMDANVPVAREEMIRYCAPAQWFARTLRRPFT
MSMEG_2563|M.smegmatis_MC2_155 HGLWELTRHPEQLAAVRTDLDANVPIAREEMIRFCAPAQWFARTVRKPFT
TH_3299|M.thermoresistible__bu HGLWELADRPDQLAAVRADPDTAVPQAREEMIRYCAPAQWFARTLRKPYT
MMAR_0399|M.marinum_M HGLWELSRRPDQLAAVRTDPQSNVNVAREEMIRYCAPAQWFARTARKPFT
MUL_1049|M.ulcerans_Agy99 RGLWELSRRPDQLAAVRTDPQSNVNVAREEMIRYCASAQWFARTARKPFT
MAV_5123|M.avium_104 QGLWELSRHPDQLAAVRADPQHNIPVAREEMIRYCAPAQWFARTVRKPFD
Mb0789c|M.bovis_AF2122/97 NAVYWAAHHPGQLARVFADHS-RIPMWVEETLRYDTSSQILARTVAHDLT
Rv0766c|M.tuberculosis_H37Rv NAVYWAAHHPGQLARVFADHS-RIPMWVEETLRYDTSSQILARTVAHDLT
MAB_1216c|M.abscessus_ATCC_199 NAVYWGHRNPDQLATVHADHD-RIPLWVEESLRYDTSSQILARTVAEDIT
MLBr_02088|M.leprae_Br4923 NAVLAMLRNPSQWKALSSNPQ-RAPLVVEETLRYDPAIHLIGRVAAKDMT
..: .* * : :: . ** :*: .. : :.*. .
Mflv_4107|M.gilvum_PYR-GCK LHGTTMRPGQRIISLLASANRDEREYPQPDEFLWNRPI-ARLLAFGRGQH
Mvan_2239|M.vanbaalenii_PYR-1 LHDTTMRPGQRIISLLASANRDEREFPDADRFVWNRPM-PRLLAFGRGQH
MSMEG_2563|M.smegmatis_MC2_155 LHGTTLQPGQRVITLLASAGRDEREYPDPDSFIWDRRI-ERLLAFGRGQH
TH_3299|M.thermoresistible__bu LHGVTMRPGQRIITLLASANRDEREFDRPDEFRWNRPI-PRTLAFGRGQH
MMAR_0399|M.marinum_M IGDQTIEPGQRVITLLASANRDEREYPDPDEFIWDRPI-RRSLAFGRGQH
MUL_1049|M.ulcerans_Agy99 IGDQTIEPGQRVITLLASANRDEREYPDPDEFIWDRPI-RRSLAFGRGQH
MAV_5123|M.avium_104 IHGQTLNPGQRVITLLASANRDEREYPDPDDFVWDRPI-RRSLAFGRGQH
Mb0789c|M.bovis_AF2122/97 LYDTTIPEGEVLLLLPGSANRDDRVFDDPDDYRIGREIGCKLVSFGSGAH
Rv0766c|M.tuberculosis_H37Rv LYDTTIPEGEVLLLLPGSANRDDRVFDDPDDYRIGREIGCKLVSFGSGAH
MAB_1216c|M.abscessus_ATCC_199 VYDTKIPAGDILLLLPGSANRDDRVFDDADQYRIGREIGAKLVSFGSGAH
MLBr_02088|M.leprae_Br4923 IGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPS-SRHLAFAVGSH
: .: *: :: * .:*.** : .* : .* : ::*. * *
Mflv_4107|M.gilvum_PYR-GCK FCLGAHLARLEITIMVTEWLKRVPEFRVDAAAASRPPSSFQWGWNNLPVD
Mvan_2239|M.vanbaalenii_PYR-1 FCLGAHLARLEIAIMVTEWLKRVPEFRVDTAAAFRPPSSFQWGWSELPVE
MSMEG_2563|M.smegmatis_MC2_155 FCLGVHLARLEITIMVTEWLKRVGEFRVDERAARRPPSSFQWGWNNVPVE
TH_3299|M.thermoresistible__bu FCLGFHLARLEIDVMVTEWLRRVPEYRIRAEDAYRPPSSFQWGWSVVPVE
MMAR_0399|M.marinum_M FCIGYHLARLEISVLLQEWFRRVPEFQICAAQATRLPSSFQWGWNKIPVE
MUL_1049|M.ulcerans_Agy99 FCIGYHLARLEIAVLLQEWFRRVPEFQICAAQATRLPSSFQWGWNKIPVE
MAV_5123|M.avium_104 FCIGYHLARLEVAVLLQEWLRRVPDYAIRADAATRLPSSFQWGWNKIPVE
Mb0789c|M.bovis_AF2122/97 FCLGAHLARMEARVALGALLRRIRNYEVDDDNVVRVHSSNVRGFAHLPIS
Rv0766c|M.tuberculosis_H37Rv FCLGAHLARMEARVALGALLRRIRNYEVDDDNVVRVHSSNVRGFAHLPIS
MAB_1216c|M.abscessus_ATCC_199 FCLGAHLARMEAKVALTELFTRISGYEIDERNSVRVHSSNVRGFAHLPMT
MLBr_02088|M.leprae_Br4923 FCLGAALARLEATVTLSAISARFPQVQLAG-ELVYKPNVAMRGMSALPVQ
**:* ***:* : : *. : . * :*:
Mflv_4107|M.gilvum_PYR-GCK VHGVGG-
Mvan_2239|M.vanbaalenii_PYR-1 V------
MSMEG_2563|M.smegmatis_MC2_155 V------
TH_3299|M.thermoresistible__bu V------
MMAR_0399|M.marinum_M V------
MUL_1049|M.ulcerans_Agy99 V------
MAV_5123|M.avium_104 V------
Mb0789c|M.bovis_AF2122/97 VQAR---
Rv0766c|M.tuberculosis_H37Rv VQAR---
MAB_1216c|M.abscessus_ATCC_199 VQLREGR
MLBr_02088|M.leprae_Br4923 V------
*