For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTDPERLPYLAVDVDNHYYEPLDAFTRHLPKEFRSRGVQMLTDGKRTYAVFGGVVNHFIPNPSFDPIIE PGCLDLLFRGEVPEGVDPASLMKVDRLTDHPEYQNRDARVAVLDRQRLETVFMLPTFACGVEEGLKHDIG ATMASIHAFNLWLDEDWGFDRPDGRIIAAPIISLADPEKAVGEVEFVLGRGAKLVCVRPAPVPGLVRPRS LGDPVHDPVWARLAEAGVPVVFHLSDSGYMAIPALWGGSGVFKGFGKRDPLDMVIMDDRAIHDTIASMIV HQVFTRHPKLKVVSIENGSYFVYRLIKRLKKSANNAPYHYKEDPVEQLKNHVWIAPYYEDDVKLLADTIG VDKILFGSDWPHGEGLADPTSFTADIPQFPEFSHEDTRRVMRHNALELLGDPDRTAPGARHLAASV
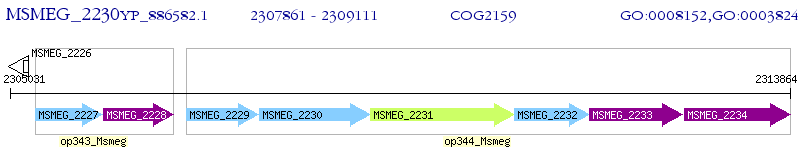
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2230 | - | - | 100% (416) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_2994 | - | 2e-73 | 38.93% (393) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4773 | - | 2e-71 | 35.92% (387) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4350 | - | 0.0 | 90.86% (394) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0299 | - | 4e-07 | 25.73% (206) | putative amidohydrolase (aminocarboxymuconate-semialdehyde |
| M. marinum M | MMAR_4014 | - | 1e-157 | 67.17% (396) | metal-dependent hydrolase |
| M. avium 104 | MAV_1591 | - | 0.0 | 77.35% (393) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_4037 | - | 0.0 | 85.50% (393) | PUTATIVE amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3878 | - | 1e-157 | 66.92% (396) | metal-dependent hydrolase |
| M. vanbaalenii PYR-1 | Mvan_1994 | - | 0.0 | 86.36% (396) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2230|M.smegmatis_MC2_155 MTTDPE----------RLPYLAVDVDNHYYEPLDAFTRHLPKEFRSRGVQ
Mflv_4350|M.gilvum_PYR-GCK MTIQQEERV----AAQRLDYRAIDVDNHYYEPIDSFTRHLPKEFRSRGVQ
Mvan_1994|M.vanbaalenii_PYR-1 MTATQQEAQRTAPAPKPLGYRAIDVDNHYYEPIDSFTRYLPKEFSRRGVQ
TH_4037|M.thermoresistible__bu -VIMTE-----------LDYRAIDVDNHYYEPLDAFTRYLPKEFTRRGVQ
MAV_1591|M.avium_104 MT--------------ALNYRVIDVDNHYYEPLDSFTRHLDKKFKRRGVQ
MMAR_4014|M.marinum_M MDS--------------LGYKAIDVDNHYYEPLDAFTRHLDKKFRRRGVQ
MUL_3878|M.ulcerans_Agy99 MDS--------------LGYKAIDVDNHYYEPLDAFTRHLDKKFRRRGVQ
MAB_0299|M.abscessus_ATCC_1997 -----------------MTSVRIDIHAHLWS-------------------
: :*:. * :.
MSMEG_2230|M.smegmatis_MC2_155 MLTDGKRTYAVFGGVVNHFIPNPSFDPIIEPGCLDLLFRGEVPEGVDPAS
Mflv_4350|M.gilvum_PYR-GCK MLNDGKRTYAVFGGVINHFIPNPTFDPIIEPGCLDLLFRGEIPEGVDPAS
Mvan_1994|M.vanbaalenii_PYR-1 MYQDGKRTWAVMGGRVNTFIPNPTFDPIIEPGCLDLLFRGEIPEGVDPAS
TH_4037|M.thermoresistible__bu MLTDGKRTFALMGEKVNHFIPNPTFDPIIEPGCLDLLFRGEIPEGVDPGS
MAV_1591|M.avium_104 MLSDGKRTWAVIGDRVNHFIPNPTFDPIIVPGCLDLLFRGEIPDGVDPAS
MMAR_4014|M.marinum_M MVTDGRHTQVIIGDRVNRFIPNPTFDPIIVPGCLDLLFRGEIPDGVDPAS
MUL_3878|M.ulcerans_Agy99 LVTDGRHTQVIIGDRVNRFIPNPTFDPIIVPGCLDLLFRGEIPDGVDPAS
MAB_0299|M.abscessus_ATCC_1997 ----------------DEYLS--------------------LLDGYGRPA
: ::. : :* . :
MSMEG_2230|M.smegmatis_MC2_155 LMKVDRLTDHPEYQNRDARVAVLDRQRLETVFMLPTFACGVEEGLKHDIG
Mflv_4350|M.gilvum_PYR-GCK LMKVDRLSEHPEYQNRDARVKILDKQNLETVFMLPTFACGVEEGLKHDIP
Mvan_1994|M.vanbaalenii_PYR-1 LMKVDRISDHPEYRNRDARVKILDKQNLETVFMLPTFACGVEEALKHDVE
TH_4037|M.thermoresistible__bu LMKVDRLANHPEYQNRDERVKILDKQNLETVFMLPTFACGVEEGLKHDIE
MAV_1591|M.avium_104 LMKVERLADHPEYQNRDARIAVMDEQDIETAFMLPTFGCGVEEALKHDIE
MMAR_4014|M.marinum_M LMKVEKLELRLEYRNRDARVAVVESQGIETIFMFPTFGCGVEEALKADIE
MUL_3878|M.ulcerans_Agy99 LMKVEKLELRPEYRNRDARVVVVESQGIETIFMFPTFGCGVEEALKADIE
MAB_0299|M.abscessus_ATCC_1997 TDVHRGLGAGATRDDLDGRFELMDEAGVDLQILTATPAS-------PHFE
: : * *. ::: :: :: .* .. ..
MSMEG_2230|M.smegmatis_MC2_155 ATMASIHAFNLWLDEDWGFDRPD-GRIIAAPIISLADPEKAVGEVEFVLG
Mflv_4350|M.gilvum_PYR-GCK ATMASVHAFNLWLDEDWGFNRPD-GRILSAPIISLADVQKAVEEVEFVLS
Mvan_1994|M.vanbaalenii_PYR-1 ATMVSVHAFNLWLDEDWGFNRPD-HRILAAPIISLADPDKAVEEAEFVLS
TH_4037|M.thermoresistible__bu ATMVSLHAFNLWLDEDWGFDRPD-GRILSAPMISLADPEKAVEEVEFVLN
MAV_1591|M.avium_104 ATMASVHAFNLWLDEDWGFDRPD-HRIIAAPIVSLADPTRAVEEVDFVLA
MMAR_4014|M.marinum_M ATAASLHAFNLWLDEDWGFDRPD-HRIIAAPMISLADPEKALQEVEFVLE
MUL_3878|M.ulcerans_Agy99 ATAASLHAFNLWLDEDWGFDRPD-HRIIAAPMISLADPEKALQEVEFVLE
MAB_0299|M.abscessus_ATCC_1997 DQAAAVASARFINDEYAKLSSEYPGRFGALASLPLPHVPAALEELARAID
.:: : .: ** :. *: : . :.*.. *: * .:
MSMEG_2230|M.smegmatis_MC2_155 RGAKLVCVRPAPVPGLVRPRSLGDPVHDPVWARLAEAGVPVVFHLSDSGY
Mflv_4350|M.gilvum_PYR-GCK RGAKIVCVRPAPVPGEVRPRSLGDPVHDPVWARLAEAGVPVVFHLSDSGY
Mvan_1994|M.vanbaalenii_PYR-1 RGAKVVCVRPAPVPGVVKPRSLGDPVHDPVWARLAEAGVPVVFHLSDSGY
TH_4037|M.thermoresistible__bu RGARIVCVRPAPVPGVVKPRSLGDPVHDPVWARLAEAGVPVIFHLSDSGY
MAV_1591|M.avium_104 RGAKLVLVRPAPVPGLVKPRSLGDRSHDPVWARLAEAGVPVGFHLSDSGY
MMAR_4014|M.marinum_M RGARMVHVRPAPVPGVPRNRSLGHRSHDRVWARLAEANVPVAFHLGDSGY
MUL_3878|M.ulcerans_Agy99 RGARMVHVRPAPVPGVPRNRSLGHRSHDRVWARLAEANVPVAFHLGDSGY
MAB_0299|M.abscessus_ATCC_1997 ELG----MYGASITTSVLGRSIADPIFDPLYAELNRRRAVLFVHPAGCG-
. . : *.:. **:.. .* ::*.* . . : .* ...*
MSMEG_2230|M.smegmatis_MC2_155 MAIPALWGGSGVFKGFGKR-DPLDMVIMDDRAIHDTIASMIVHQVFTRHP
Mflv_4350|M.gilvum_PYR-GCK TAIPALWGGSGVFKGFGKR-DPLDMVIMDDRAIHDTIASMIVHQVFTRHP
Mvan_1994|M.vanbaalenii_PYR-1 MAIPALWGGKDTFEGFGKR-DPLDMVVMDDRAIHDSIASMIVHQVFTRHP
TH_4037|M.thermoresistible__bu LAISALWGGTGTFEGFGKR-DPLDNVLLDDRAIHDTMASMIVHQVFTRHP
MAV_1591|M.avium_104 LHIAAAWGGKSTFEGFGAK-DPLDQVLLDDRAIHDTMASMIVHGVFTRHP
MMAR_4014|M.marinum_M LEIAGMWGGKETFEPFAAPPDPLDKILVDDRAIHDTMASLIVHGVFDRHP
MUL_3878|M.ulcerans_Agy99 LEIAGMWGGKETFEPFAAPPDPLDKILVDDRAIHDTMASLIVHGVFDRHP
MAB_0299|M.abscessus_ATCC_1997 -AESSLITDHSLTWSIGAP--------IED---TVAIMHLIVAGVPSRYP
.. . :. ::* :: :** * *:*
MSMEG_2230|M.smegmatis_MC2_155 KLKVVSIENGSYFVYRLIKRLKKSANNAPYHYKEDPVEQLKNHVWIAP--
Mflv_4350|M.gilvum_PYR-GCK KLKVASIENGSYFVYRLIKRLKKSANNAPYHYKEDPVEQLRNNVWIAP--
Mvan_1994|M.vanbaalenii_PYR-1 TLKVASIENGSYFVYRLIKRLKKSANNAPYHYKEDPVEQLRNNVWIAP--
TH_4037|M.thermoresistible__bu NLKVASIENGSYFVYRLIKRLKKAANTAPSHFKEDPVEQLKNNVWIAP--
MAV_1591|M.avium_104 KLKAVSIENGSYFVHRLIKRLKKAANTQPQYFPEDPVEQLRNNVWIAP--
MMAR_4014|M.marinum_M TLRVASIENGSEWVHRLAKRLKKLANQHPRSFPNDPVDALREHVWISP--
MUL_3878|M.ulcerans_Agy99 TLRVASIENGSEWVHRLAKRLKKLANQHPRSFPNDPVDALREHVWISP--
MAB_0299|M.abscessus_ATCC_1997 DMKIVTCHLGGALPMVLERAHRQVTEWEATQCPEAPRDAARRLWYDTVGH
:: .: . *. * : :: :: . : * : :. : :
MSMEG_2230|M.smegmatis_MC2_155 YYEDDVKLLADTIGVDKILFGSDWPHGEGLADPTSFTADIPQFPEFSHED
Mflv_4350|M.gilvum_PYR-GCK YYEDDVKLLAETIGVDKILFGSDWPHGEGLADPTSFTADIPQFPEFSHED
Mvan_1994|M.vanbaalenii_PYR-1 YYEDDVKLLADTIGVDKILFGSDWPHGEGLADPTSFTADIPQFPEFSHED
TH_4037|M.thermoresistible__bu YYEDDVKLLAETIGVDRILFGSDWPHGEGLADPMTFTADIPQFPEFSHED
MAV_1591|M.avium_104 YYEDDLPELARVIGVDKILFGSDWPHGEGLASPVSFTAELKGFSES---D
MMAR_4014|M.marinum_M YYEEDLTVLADRIGVDRILFGSDWPHGEGLESPLAFTEELTGFSRA---E
MUL_3878|M.ulcerans_Agy99 YYEEDLTVLADRIGVDRILFGSDWPHGEGLESPLAFTLELTGFSRA---A
MAB_0299|M.abscessus_ATCC_1997 DHTPALRAAVASLGVDRLIFGSDFPYQAGPRYVSSATYIEEGLSAE---D
: : . :***:::****:*: * : * :.
MSMEG_2230|M.smegmatis_MC2_155 TRRVMRHNALELLGDPDRTAPGARHLAASV
Mflv_4350|M.gilvum_PYR-GCK TRKVMRDNALTLVHGA--------------
Mvan_1994|M.vanbaalenii_PYR-1 TRKVMRDNALTLVGTHVSA-----------
TH_4037|M.thermoresistible__bu TRKVMRGNALTLLGANVPATA---------
MAV_1591|M.avium_104 IRKIMRDNALDLLGVQVGSAA---------
MMAR_4014|M.marinum_M VQKIMRDNALDFLGVTVASAA---------
MUL_3878|M.ulcerans_Agy99 VQKIMRDNALDFLGVTVASVA---------
MAB_0299|M.abscessus_ATCC_1997 ASAILDHNGAELLGLTGV------------
:: *. ::