For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIVLTADHGAVRVLTMNRPEARNALGSELIEALFTALQDADDDESVRAVVLTGADPAFCAGVDLKQAQTL GMDYFARFQTHNCIAKTGQMATPIIGAINGATFTGGLEMALGCDFLIASERAVFADTHARVGILPGGGMT ARLPQVVGAAMARRLSMTGEVVDASRAERLGLVTEVVAHEQLLVRALELAQQIAEVPGPIMSGLKEIYTL GADPVITPALDAEQQIAGTQQRDFAGLGDRYRAVAERNHAQIQR*
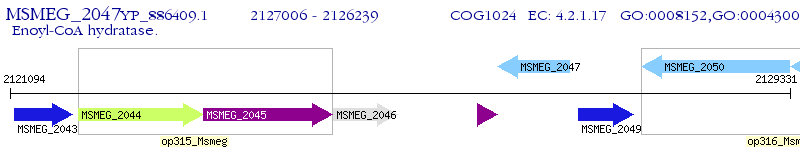
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2047 | - | - | 100% (255) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_2641 | - | 5e-49 | 44.88% (254) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_4020 | - | 1e-32 | 35.39% (243) | enoyl-CoA hydratase/isomerase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2855 | echA16 | 4e-49 | 53.61% (194) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_4495 | - | 1e-103 | 74.10% (251) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv2831 | echA16 | 5e-49 | 53.61% (194) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 4e-32 | 32.59% (224) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3124 | - | 1e-46 | 45.10% (255) | enoyl-CoA hydratase |
| M. marinum M | MMAR_1903 | echA16_2 | 3e-46 | 52.43% (185) | enoyl-CoA hydratase, EchA16_2 |
| M. avium 104 | MAV_3689 | - | 3e-47 | 52.60% (192) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3880 | - | 1e-102 | 72.91% (251) | PUTATIVE Enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 1e-31 | 35.94% (192) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_1871 | - | 1e-100 | 73.81% (252) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2047|M.smegmatis_MC2_155 MS---IVLTADHGAVRVLTMNRPEARNALGSELIEALFTALQDADDDESV
TH_3880|M.thermoresistible__bu MTSDRVVSRTDHGPVRLLTMNRPESRNALSRNLIRELYTAFEEADQDASV
Mflv_4495|M.gilvum_PYR-GCK MT-EDLVLTADHDGVRVITLNRPAARNALSRDLIRATYTALTSADDDDAV
Mvan_1871|M.vanbaalenii_PYR-1 MT-AELVLTADHDGVRVITLNRPAARNALSRDLIRASYAALTAADADESV
Mb2855|M.bovis_AF2122/97 MT-DDILLIDTDERVRTLTLNRPQSRNALSAALRDRFFAALADAEADDDI
Rv2831|M.tuberculosis_H37Rv MT-DDILLIDTDERVRTLTLNRPQSRNALSAALRDRFFAALADAEADDDI
MMAR_1903|M.marinum_M MT-NEILLIDTAERVLTLTLNRPQSRNALSAALRDQFFAALAAAETDDDV
MAV_3689|M.avium_104 MT-DEILLSNTEERVRTLTLNRPQARNALSAALRDRFFGALADAETDDDV
MAB_3124|M.abscessus_ATCC_1997 MT-EPILLTHTEDRICTITLNRPQARNALSTALGEEIVKAVTAADADENV
MLBr_02402|M.leprae_Br4923 MK-YDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELDIDPDV
MUL_0210|M.ulcerans_Agy99 MS-YETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELDNDPGI
*. : : :*:*** : ***. : * : * :
MSMEG_2047|M.smegmatis_MC2_155 RAVVLTGADPAFCAGVDLKQAQTLGMDYFARFQTHNCIAKTGQMATPIIG
TH_3880|M.thermoresistible__bu RAMVLTGADPAFCAGVDLKEAQRDGMTYFDEFRTHDCITKPAELRTPIIA
Mflv_4495|M.gilvum_PYR-GCK RAVVLTGADPAFCAGVDLKEAARDGLSYFAEFRTQSCIAAVAAMRTPVVG
Mvan_1871|M.vanbaalenii_PYR-1 RAVVLTGADPAFCAGVDLKEAARDGLSYFAEFRSQSCIAAVARMRTPVVG
Mb2855|M.bovis_AF2122/97 DVVILTGADPVFCAGLDLKELA--GQTALPDISP-----RWPAMTKPVIG
Rv2831|M.tuberculosis_H37Rv DVVILTGADPVFCAGLDLKELA--GQTALPDISP-----RWPAMTKPVIG
MMAR_1903|M.marinum_M DVIILTGADPVFCAGLDLKELG--GQSALPDISP-----RWPALTKPVIG
MAV_3689|M.avium_104 DVVIITGADPVFCAGLDLKELG--GSSALPDISP-----RWPALTKPVIG
MAB_3124|M.abscessus_ATCC_1997 DVMILTAADPVFCAGVDLKELG--SGDRAESLDP-----WWPELSKPVIG
MLBr_02402|M.leprae_Br4923 GAILITGSPKVFAAGADIKEMASLTFTDAFDADFFSAWGKLAAVRTPMIA
MUL_0210|M.ulcerans_Agy99 GAIIITGSAKAFAAGADIKEMADLTFSEAFDADFFAAWGKLAAVRTPTIA
.:::*.: .*.** *:*: : .* :.
MSMEG_2047|M.smegmatis_MC2_155 AINGATFTGGLEMALGCDFLIASERAVFADTHARVGILPGGGMTARLPQV
TH_3880|M.thermoresistible__bu AVNGPTFTGGLEMALGCDFLIASDRAVFADTHARVGILPGGGMTARLPQV
Mflv_4495|M.gilvum_PYR-GCK AINGATFTGGLEMALGCDFMIASERAVFADTHARVGILPGGGMTARLPQL
Mvan_1871|M.vanbaalenii_PYR-1 AINGATFTGGLEMALGCDFLIASERAVFADTHARVGILPGGGMTAYLPQV
Mb2855|M.bovis_AF2122/97 AINGAAVTGGLELALYCDILIASEHARFADTHARVGLLPTWGLSVRLPQK
Rv2831|M.tuberculosis_H37Rv AINGAAVTGGLELALYCDILIASEHARFADTHARVGLLPTWGLSVRLPQK
MMAR_1903|M.marinum_M AINGAAVTGGLELALYCDILIASEQARFADTHARVGLLPTWGLSVRLPQK
MAV_3689|M.avium_104 AINGAAVTGGLELALYCDILIASENARFADTHARVGLLPTWGLSVRLPQK
MAB_3124|M.abscessus_ATCC_1997 AINGAAVTGGLELALACDILIASEKARFADTHARVGILPTWGLTTLLPLS
MLBr_02402|M.leprae_Br4923 AVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRA
MUL_0210|M.ulcerans_Agy99 AVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQRLTRA
*: * :. ** *:*: **::**:: * *.:.. ::*:** * : *.
MSMEG_2047|M.smegmatis_MC2_155 VGAAMARRLSMTGEVVDASRAERLGLVTEVVAHEQLLVRALELAQQIAEV
TH_3880|M.thermoresistible__bu VGAAMARRMSMTGEVIDAARAERLGLVTEVVEHDRLLDRALELAGQIAEV
Mflv_4495|M.gilvum_PYR-GCK VGAAMARRLSMTGEVVDAARAERIGLVTEVVAHQHLLQRAIDLAHQIAEV
Mvan_1871|M.vanbaalenii_PYR-1 VGAAMARRLSMTGEVVDAARAERIGLVTEVVVHDRLLDRAVELATQIAEV
Mb2855|M.bovis_AF2122/97 VGIGLARRMSLTGDYLSATDALRAGLVTEVVAHDQLLPTARRVAASIVGN
Rv2831|M.tuberculosis_H37Rv VGIGLARRMSLTGDYLSATDALRAGLVTEVVAHDQLLPTARRVAASIVGN
MMAR_1903|M.marinum_M VGVGLARRMSLTGDYLSATDALRAGLVTEVVEHQQLLPAARQVAASIVGN
MAV_3689|M.avium_104 VGIGLARRMSLTGDYLSAADALRAGLVTEVVPHDQLLGAARAVAASIVGN
MAB_3124|M.abscessus_ATCC_1997 VGRGLARRMSLTGDYLSAEDALRAGLVTEVVAHNELLPAAKRLAATIAGN
MLBr_02402|M.leprae_Br4923 IGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQM
MUL_0210|M.ulcerans_Agy99 IGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVATTISQM
:* . * : :** :.* * * ***:.** : ** * :* *
MSMEG_2047|M.smegmatis_MC2_155 PGPIMSGLKEIYTLGADPVITPALDAEQQIAGTQQRD---FAGLGDRYRA
TH_3880|M.thermoresistible__bu PADTMAGLKEIYARGIGPVIAPALAAEQDIAAAQLRD---FGGLGERFMA
Mflv_4495|M.gilvum_PYR-GCK PRPTMRGLKEIYTAGASAVVDPALAAEERIAFAQHRD---LEGLGDRFSA
Mvan_1871|M.vanbaalenii_PYR-1 PRPTMRSLKEIYTTGAAAVTDPALAAEEEIAFAQHRD---LDGLGDRFSA
Mb2855|M.bovis_AF2122/97 NQNAVRALLASYHRIDESQTAAGLWLEACAAKQFCTS---GDTIAANREA
Rv2831|M.tuberculosis_H37Rv NQNAVRALLASYHRIDESQTAAGLWLEACAAKQFRTS---GDTIAANREA
MMAR_1903|M.marinum_M NQNAVRALLASYHRIDESSTAQGLWLEATAAREFRTS---GDDIAANREA
MAV_3689|M.avium_104 NQNAVRALLTSYHRIDDAQTSAGLWQEAMAARQFRTS---GDDIAANREA
MAB_3124|M.abscessus_ATCC_1997 NQPAVRELLASYRRIESELVGNGLQVALDDAHRWIDQNSIAEGVEARRTA
MLBr_02402|M.leprae_Br4923 SRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVT----DDQSEGMAA
MUL_0210|M.ulcerans_Agy99 SRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFAT----ADQSEGMAA
.* *
MSMEG_2047|M.smegmatis_MC2_155 VAERNHAQIQR-
TH_3880|M.thermoresistible__bu VSQRNRQQLG--
Mflv_4495|M.gilvum_PYR-GCK VSQRNRDQIGGA
Mvan_1871|M.vanbaalenii_PYR-1 VAQRNKEQIRPS
Mb2855|M.bovis_AF2122/97 VLQRGRAQVR--
Rv2831|M.tuberculosis_H37Rv VLQRGRAQVR--
MMAR_1903|M.marinum_M VLNRGRSQVR--
MAV_3689|M.avium_104 VLARGRSQVR--
MAB_3124|M.abscessus_ATCC_1997 IMERGRSQNA--
MLBr_02402|M.leprae_Br4923 FIEKRAPQFTHR
MUL_0210|M.ulcerans_Agy99 FIEKRPPNFTHR
. : :