For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTQIQHFINGRRTSAGSTRTADVMNPSTGEVQAQVLLASAADVDAAVTGAAEAQKEWAAYNPQRRARVLM KFIDLVNQNADELARLLSIEHGKTVADSLGDIQRGIEVIEFAVGIPHLLKGEFTEGAGTGIDVYSIRQPL GVVAGITPFNFPAMIPLWKAGPALACGNAFILKPSERDPSVPVRLAELFIEAGLPPGVFQVVQGDKEAVD AILTHPTVQAVGFVGSSDIAQYIYSTAAAHGKRSQCFGGAKNHMIIMPDADLDQAVDALIGAGYGSAGER CMAISVAVPVGEETANRLRARLTERVKQLRVGHSLDPKADYGPLVTEAALNRVKDYIGQGVEAGAELVVD GRERATDELTFDDQSLEGGYFIGPTLFDHVKTDMSIYTDEIFGPVLCIVRAHDYEEALSLPTKHEYGNGV AIFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPASIQFYTKTKTVTERWP SGIKDGAEFVIPTMK*
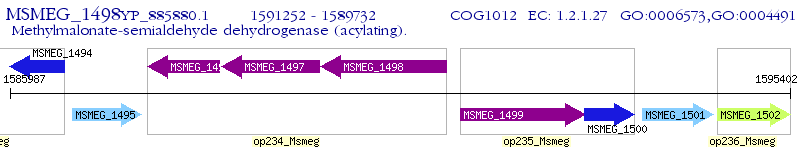
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1498 | mmsA | - | 100% (506) | methylmalonate-semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2449 | mmsA | e-148 | 51.98% (506) | methylmalonate-semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6687 | - | 5e-75 | 34.02% (482) | aldehyde dehydrogenase, thermostable |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0775c | mmsA | 0.0 | 83.10% (509) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5010 | - | 0.0 | 90.91% (506) | methylmalonate-semialdehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0753c | mmsA | 0.0 | 83.30% (509) | methylmalonate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 9e-48 | 29.03% (465) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1189c | - | 0.0 | 71.91% (502) | methylmalonic acid semialdehyde dehydrogenase MmsA |
| M. marinum M | MMAR_1079 | mmsA | 0.0 | 86.17% (506) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. avium 104 | MAV_4417 | mmsA | 0.0 | 85.15% (505) | methylmalonate-semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_2909 | - | 0.0 | 86.17% (506) | PUTATIVE methylmalonate-semialdehyde dehydrogenase [acylating) |
| M. ulcerans Agy99 | MUL_0837 | mmsA | 0.0 | 85.97% (506) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. vanbaalenii PYR-1 | Mvan_1362 | - | 0.0 | 89.72% (506) | methylmalonate-semialdehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5010|M.gilvum_PYR-GCK ----------------------------MTTRIPHFIDGKRSELASTRTA
Mvan_1362|M.vanbaalenii_PYR-1 MTHTTRAPCQGRVRITRISKYPPLEGSLMTTRIPHFIDGKRSDLSSTRTA
MSMEG_1498|M.smegmatis_MC2_155 ----------------------------MTTQIQHFINGRRTSAGSTRTA
Mb0775c|M.bovis_AF2122/97 ----------------------------MTTQISHFIDGQRTAGQSTRSA
Rv0753c|M.tuberculosis_H37Rv ----------------------------MTTQISHFIDGQRTAGQSTRSA
MAV_4417|M.avium_104 ----------------------------MTTQIPHFIDGQRTAGQSTRTA
MMAR_1079|M.marinum_M ----------------------------MTTQIPHFIDGRRTPGQSTRTA
MUL_0837|M.ulcerans_Agy99 ----------------------------MTTQIPHFIDGRRTPGQSTRTA
TH_2909|M.thermoresistible__bu ----------------------------MTRQIHHFINGQHTAGQSTRTA
MAB_1189c|M.abscessus_ATCC_199 --------------------------MSSLPQLTHFVAGKPVPGTSGRFA
MLBr_02573|M.leprae_Br4923 -----------------------------------------------MSI
Mflv_5010|M.gilvum_PYR-GCK GVLNPSTGEVQSEVLLASAADVDTAVASAVEAQKEWAAWNPQRRARVFMK
Mvan_1362|M.vanbaalenii_PYR-1 DVLNPSTGAVQAEVLLASAADVDTAVASAVEAQKEWAAWNPQRRARVMMK
MSMEG_1498|M.smegmatis_MC2_155 DVMNPSTGEVQAQVLLASAADVDAAVTGAAEAQKEWAAYNPQRRARVLMK
Mb0775c|M.bovis_AF2122/97 DVFDPNTGQIQAKVPMAGKSDIDAAVASAVEAQKGWAAWNPQRRARVLMR
Rv0753c|M.tuberculosis_H37Rv DVFDPNTGQIQAKVPMAGKSDIDAAVASAVEAQKGWAAWNPQRRARVLMR
MAV_4417|M.avium_104 EVFDPSTGAVQAQVPMAGRADIDAAVASAIEAQKGWAATNPQRRARVLMR
MMAR_1079|M.marinum_M DVLDPNTGQVQATVPMAAQADIDAAVAGAAEAQIGWAAWNPQRRARVLMR
MUL_0837|M.ulcerans_Agy99 DVLDPNTGQVQATVPMAAQADIDAAVAGAAEAQVGWAAWNPQRRARVLMR
TH_2909|M.thermoresistible__bu DVMNPSTGKVQAKVVMGTAADIDAAVAGAAEAQKEWAAWNPQRRARVLMK
MAB_1189c|M.abscessus_ATCC_199 DVYDPTQGKPVRQVPLADVSEVEAAISNAAEAQPGWAARNPQQRARVLAK
MLBr_02573|M.leprae_Br4923 ATINPATGETVKTFTPTTQDEVEDAIARAYARFDNYRHTTFTQRAKWANA
. :* * . ::: *:: * : . :**:
Mflv_5010|M.gilvum_PYR-GCK FIQLVNDHVDELAELLSIEHGKTVADSKGDIQRGIEVIEFAIG-IPHLLK
Mvan_1362|M.vanbaalenii_PYR-1 FIELVNANIDELAELLSIEHGKTVPDAKGDIQRGIEVIEFAIG-IPHLLK
MSMEG_1498|M.smegmatis_MC2_155 FIDLVNQNADELARLLSIEHGKTVADSLGDIQRGIEVIEFAVG-IPHLLK
Mb0775c|M.bovis_AF2122/97 FIELVNDTIDELAELLSREHGKTLADARGDVQRGIEVIEFCLG-IPHLLK
Rv0753c|M.tuberculosis_H37Rv FIELVNDTIDELAELLSREHGKTLADARGDVQRGIEVIEFCLG-IPHLLK
MAV_4417|M.avium_104 FIELVNQHNDELAELLSREHGKTLADAKGDIQRGIEVIEFCVG-IPHLLK
MMAR_1079|M.marinum_M FIELVNQNTDELAELLSREHGKTVPDARGDIQRGIEVIEFCLG-IPHLLK
MUL_0837|M.ulcerans_Agy99 FIELVNQNTDELAELLSREHGKTVPDARGDIQRGIEVIEFCLG-IPHLLK
TH_2909|M.thermoresistible__bu FIDLVNQNVDELAELLSLEHGKTLADAKGDIQRGIEVIEFSIG-IPHLQK
MAB_1189c|M.abscessus_ATCC_199 FVDLVRAEIDDLAAMLSQEHGKTLADAKGDIERGLEVCEFATG-IPHLIK
MLBr_02573|M.leprae_Br4923 TADLLEAEANQSAALMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAETLLA
:*:. :: * ::: * ***: .: .: : : .: *
Mflv_5010|M.gilvum_PYR-GCK GEFTENAGTGIDVYSIR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
Mvan_1362|M.vanbaalenii_PYR-1 GEFTEGAGSGIDVYSIR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MSMEG_1498|M.smegmatis_MC2_155 GEFTEGAGTGIDVYSIR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
Mb0775c|M.bovis_AF2122/97 GEYTEGAGPGIDVYSLR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
Rv0753c|M.tuberculosis_H37Rv GEYTEGAGPGIDVYSLR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MAV_4417|M.avium_104 GEYSEGAGPGIDVYSLR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MMAR_1079|M.marinum_M GEYTEGAGPGIDVYSLR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MUL_0837|M.ulcerans_Agy99 GEYTEGAGPGIDVYSLR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
TH_2909|M.thermoresistible__bu GEYTEGAGPGIDVYSMR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MAB_1189c|M.abscessus_ATCC_199 GEYTSGAGTGIDVYSIR-QPLGVVAGITPFNFPAMIPLWKAGPALACGNA
MLBr_02573|M.leprae_Br4923 DEPAEAAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNV
.* :. *. * . * ****** .: *:*** .: *.*** .**.
Mflv_5010|M.gilvum_PYR-GCK FILKPSERDPSVPLRLAELFLEAGLPAGVFQVVQGDKEAVDAILTHPDIQ
Mvan_1362|M.vanbaalenii_PYR-1 FILKPSERDPSVPVRLAELFLEAGLPAGVFQVVHGDKEAVDAILTHPDIQ
MSMEG_1498|M.smegmatis_MC2_155 FILKPSERDPSVPVRLAELFIEAGLPPGVFQVVQGDKEAVDAILTHPTVQ
Mb0775c|M.bovis_AF2122/97 FVLKPSERDPSVPVRLAELFIEAGLPAGVFQVVHGDKEAVDAILHHPDIK
Rv0753c|M.tuberculosis_H37Rv FVLKPSERDPSVPVRLAELFIEAGLPAGVFQVVHGDKEAVDAILHHPDIK
MAV_4417|M.avium_104 FVLKPSERDPSVPVRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPDIQ
MMAR_1079|M.marinum_M FVLKPSERDPSVPLRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPEIK
MUL_0837|M.ulcerans_Agy99 FVLKPSERDPSVPLRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPEIK
TH_2909|M.thermoresistible__bu FILKPSERDPSVPVRLAELFLQAGLPSGLFQVVHGDKEAVDAILEHPTIQ
MAB_1189c|M.abscessus_ATCC_199 FVLKPSERDPSVPLRLAELFLEAGLPPGVFNVVNGDKTAVDALLHDPRVA
MLBr_02573|M.leprae_Br4923 GLLKHASNVPQCALYLADVIARGGFPEDCFQTLLISANGVEAILRDPRVA
:** :.. *. .: **::: ..*:* . *:.: . .*:*:* .* :
Mflv_5010|M.gilvum_PYR-GCK AVGFVGSSDIAQYIYSTAAAHGKRSQCFGGAKNHMIIMPDADLDQAVDAL
Mvan_1362|M.vanbaalenii_PYR-1 AVGFVGSSDIAQYIYSTAAANGKRSQCFGGAKNHMIIMPDADLDQAVDAL
MSMEG_1498|M.smegmatis_MC2_155 AVGFVGSSDIAQYIYSTAAAHGKRSQCFGGAKNHMIIMPDADLDQAVDAL
Mb0775c|M.bovis_AF2122/97 AVGFVGSSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDAL
Rv0753c|M.tuberculosis_H37Rv AVGFVGSSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDAL
MAV_4417|M.avium_104 AVGFVGSSDIAQYIYAGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDAL
MMAR_1079|M.marinum_M AVGFVGSSDIAQYIYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDAL
MUL_0837|M.ulcerans_Agy99 AVGFVGSSDIAQYIYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDAL
TH_2909|M.thermoresistible__bu AVGFVGSSDIAQYIYAGATAHGKRAQCFGGAKNHMIVMPDADLDQAVDAL
MAB_1189c|M.abscessus_ATCC_199 AVGFVGSTPIAQYIYETATANGKRAQCFGGAKNHMIIMPDADIDQAIDAL
MLBr_02573|M.leprae_Br4923 AATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATA
*. :.** .* : * * : *..: :*:**.**:* *: :
Mflv_5010|M.gilvum_PYR-GCK IGAGYGSAGERCMAISVAVPVGEETANRLRNRLVERVNQLRVGHSLDPKA
Mvan_1362|M.vanbaalenii_PYR-1 IGAGYGSAGERCMAISVAVPVGEETANRLRNRLVERVNQLRVGHSLDPKA
MSMEG_1498|M.smegmatis_MC2_155 IGAGYGSAGERCMAISVAVPVGEETANRLRARLTERVKQLRVGHSLDPKA
Mb0775c|M.bovis_AF2122/97 IGAGYGSAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKA
Rv0753c|M.tuberculosis_H37Rv IGAGYGSAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKA
MAV_4417|M.avium_104 IGAGYGSAGERCMAISVAVPVGDKTADRLRARLVERINNLRVGHSLDPKA
MMAR_1079|M.marinum_M IGAGYGSAGERCMAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKA
MUL_0837|M.ulcerans_Agy99 IGAGYGSTGERCMAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKA
TH_2909|M.thermoresistible__bu IGAGYGSAGERCMAISVAVPVGDEIADRLRARLIERVANLRVGHSLDPKA
MAB_1189c|M.abscessus_ATCC_199 IGAGYGSAGERCMAISVAVPVGEATAERLVDGLTKRARELVVGASLDEGV
MLBr_02573|M.leprae_Br4923 VTGRVQNNGQSCIAAKRFIAH-ADIYDEFVEKFVARMETLRVGDPTDPDT
: . . *: *:* . :. :.: : * * ** . * .
Mflv_5010|M.gilvum_PYR-GCK DYGPLVTGAALERVRDYIGQGVEAGAELVVDGRERATDELSF----DDQD
Mvan_1362|M.vanbaalenii_PYR-1 DYGPLVTGAALERVRDYIGQGVDAGAELVVDGRERTTDELSF----DDQD
MSMEG_1498|M.smegmatis_MC2_155 DYGPLVTEAALNRVKDYIGQGVEAGAELVVDGRERATDELTF----DDQS
Mb0775c|M.bovis_AF2122/97 DYGPLVTGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDAN
Rv0753c|M.tuberculosis_H37Rv DYGPLVTGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDAN
MAV_4417|M.avium_104 DYGPLVTGAAVARVRDYIDQGVAAGAEIVVDGRERRSDDLTF----GDAN
MMAR_1079|M.marinum_M DYGPLVTEAALTRVRDYIGQGVASGAELVVDGRERASDDLAF----GDAN
MUL_0837|M.ulcerans_Agy99 DYGPLVTEAALTRVRDYIGQGVASGAELVVDGRERASDDLAF----GDAN
TH_2909|M.thermoresistible__bu DYGPLVTEAALQRVRDYIGQGVEAGAELLVDGRERGSDDLQF----GDDS
MAB_1189c|M.abscessus_ATCC_199 DFGPLVGADALKRVRDYIDIGVAEGAELVLDGRDLTVD-----------G
MLBr_02573|M.leprae_Br4923 DVGPLATESGRAQVEQQVDAAAAAGAVIRCGGKR---------------P
* ***. . :*.: :. .. ** : .*:
Mflv_5010|M.gilvum_PYR-GCK LSKGYFIGPTLFDHVTTDMSIYTDEIFGPVLCIVRAADYDEALSLPTKHE
Mvan_1362|M.vanbaalenii_PYR-1 LSKGFFIGPTLFDHVTTDMSIYTGEIFGPVLCIVRAKDYDEALSLPSKHE
MSMEG_1498|M.smegmatis_MC2_155 LEGGYFIGPTLFDHVKTDMSIYTDEIFGPVLCIVRAHDYEEALSLPTKHE
Mb0775c|M.bovis_AF2122/97 LEGGFFIGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHE
Rv0753c|M.tuberculosis_H37Rv LEGGFFIGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHE
MAV_4417|M.avium_104 LEGGFFIGPTLFDHVTPDMSIYTDEIFGPVLCIVRAQNYEEALRLPSEHE
MMAR_1079|M.marinum_M LEGGYFIGPTLFDHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHE
MUL_0837|M.ulcerans_Agy99 LEGGYFIGPTLFDHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHE
TH_2909|M.thermoresistible__bu LEGGYFIGPTLFDRVTTDMSIYRDEIFGPVLCMVRAKNYEEALALPSEHE
MAB_1189c|M.abscessus_ATCC_199 HKDGFFIGASLFDHVTPEMRIYKEEIFGPVVSVVRAKDYDEAVRLPSEHE
MLBr_02573|M.leprae_Br4923 DRPGWFYPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATP
*:* .::: : .* :* *:**** .: * : ::*: :..
Mflv_5010|M.gilvum_PYR-GCK YGNGVAIFTRDGDAARDFVSKVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
Mvan_1362|M.vanbaalenii_PYR-1 YGNGVAIFTRDGDAARDFVSKVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
MSMEG_1498|M.smegmatis_MC2_155 YGNGVAIFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
Mb0775c|M.bovis_AF2122/97 YGNGVAIFTRDGDAARDFVSRGQVGMVGVNVPIPVPVAYHTFGGWKRSGF
Rv0753c|M.tuberculosis_H37Rv YGNGVAIFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
MAV_4417|M.avium_104 YGNGVAVFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
MMAR_1079|M.marinum_M YGNGVAVFTRDGDTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
MUL_0837|M.ulcerans_Agy99 YGNGVAVFTRDGDTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGF
TH_2909|M.thermoresistible__bu YGNGVAIFTRDGDTARDFVSRVQCGMVGVNVPIPVPVAYHTFGGWKRSGF
MAB_1189c|M.abscessus_ATCC_199 FGNGVAIFTRDGDTARDFCAKVNTGMVGVNVPIPVPVAYHTFGGWKASGF
MLBr_02573|M.leprae_Br4923 FGLGSNAWTQNESEQRHFIDNIVAGQVFING-MTVSYPELPFGGIKRSGY
:* * :*:: . *.* . * * :* :.*. . .*** * **:
Mflv_5010|M.gilvum_PYR-GCK G-DLNQHGPASIQFYTKVKTVTERWPSGIKDGAEFVIPTMK
Mvan_1362|M.vanbaalenii_PYR-1 G-DLNQHGPASIQFYTKVKTVTERWPSGIKDGAEFVIPTMK
MSMEG_1498|M.smegmatis_MC2_155 G-DLNQHGPASIQFYTKTKTVTERWPSGIKDGAEFVIPTMK
Mb0775c|M.bovis_AF2122/97 G-DLNQHGPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS
Rv0753c|M.tuberculosis_H37Rv G-DLNQHGPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS
MAV_4417|M.avium_104 G-DLNQHGPASIQFYTKVKTVTSRWPSGIKDGAEFVIPTMD
MMAR_1079|M.marinum_M G-DLNQHGPTSIQFYTKVKTVTSRWPSGIKDGAEFVIPTMK
MUL_0837|M.ulcerans_Agy99 G-DLNQHGPTSIQFYTKVKTVTSRWPSGIKDGAEFVIPTMK
TH_2909|M.thermoresistible__bu G-DLNQHGPHSILFYTKTKTVTQRWPSGIKDGAEFSIPTMK
MAB_1189c|M.abscessus_ATCC_199 G-DLNQHGPDSIRFYTKTKTVTQRWPSGIKEGASFVIPTMD
MLBr_02573|M.leprae_Br4923 GRELSTHG---IREFCNIKTVWIA-----------------
* :*. ** * : : ***