For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEAVDALPPAVNPLALSLNEFPYPPLPAVKAALSAAVDAANRYPEFLPERLRSLIAGRVGVDEGQVIIGV GATGVIMQVLHAVTSPGDTMVMSAPTFDGYPIFAQMARLRSVTVDLDVYGHHNLHAMADAARDARVVVVC RPHNPTGTIEPAEDIERFLLESIPEDTVVLLDEAYAEFLGPANRIDGPRLVERFRNVVVVRTFSKAYGLA GLRIGYGFCAPQLARQLWTMQLPFGIGLPALVAVAASYDAEGQLRQRIRMISAERRFLQMRLRSMGVYST DGQANFVYLPATGVPWHDVFDDAGLKVRHYADGGVRVTVGTRESTREVLRATRLALR
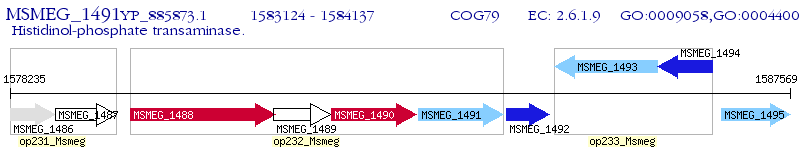
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1491 | - | - | 100% (337) | histidinol-phosphate aminotransferase 2 |
| M. smegmatis MC2 155 | MSMEG_6351 | hisC | 7e-44 | 35.81% (310) | putative aminotransferase |
| M. smegmatis MC2 155 | MSMEG_3206 | hisC | 8e-25 | 29.52% (332) | histidinol-phosphate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3800 | hisC2 | 6e-47 | 36.81% (326) | putative aminotransferase |
| M. gilvum PYR-GCK | Mflv_1195 | - | 3e-42 | 35.69% (311) | putative aminotransferase |
| M. tuberculosis H37Rv | Rv3772 | hisC2 | 2e-46 | 36.50% (326) | putative aminotransferase |
| M. leprae Br4923 | MLBr_01258 | hisC | 4e-24 | 28.86% (343) | histidinol-phosphate aminotransferase |
| M. abscessus ATCC 19977 | MAB_2168 | - | 3e-80 | 50.79% (317) | putative aminotransferase |
| M. marinum M | MMAR_5326 | hisC2 | 7e-44 | 34.24% (330) | histidinol-phosphate aminotransferase HisC2 |
| M. avium 104 | MAV_4421 | - | 1e-111 | 62.01% (329) | histidinol-phosphate aminotransferase |
| M. thermoresistible (build 8) | TH_3677 | - | 1e-114 | 62.95% (332) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_4407 | hisC2 | 8e-44 | 34.24% (330) | putative aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_1355 | - | 1e-136 | 71.94% (335) | aminotransferase, class I and II |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3800|M.bovis_AF2122/97 ---------MTARLRPELAGLP--------VYVPGKTVPGAIKLASNETV
Rv3772|M.tuberculosis_H37Rv ---------MTARLRPELAGLP--------VYVPGKTVPGAIKLASNETV
MMAR_5326|M.marinum_M ---------MTARLRPVLAGLP--------VYVPGKTVPGAIKLASNETV
MUL_4407|M.ulcerans_Agy99 ---------MTAHLRPVLAGLP--------VYVPGKTVPGAIKLASNETV
Mflv_1195|M.gilvum_PYR-GCK MRAGHKLSQVSARLRPELADLP--------AYTPGRTVPGAIKIASNETV
MSMEG_1491|M.smegmatis_MC2_155 -----------------MEAVD---------ALPPAVNP--LALSLNEFP
Mvan_1355|M.vanbaalenii_PYR-1 ----------MASRDTLAEAVT---------ALPRAVDP--MALSLNEFP
MAV_4421|M.avium_104 ---------MLTVPRPADGGIG--------EALPDAVDP--FALSLNENP
TH_3677|M.thermoresistible__bu -------MTSLIEAPPAERSLPGRRQPDTTPVVPSAVDP--MALSLNESP
MAB_2168|M.abscessus_ATCC_1997 -----------------MGREG-----------PVTVSP--LRLALNENP
MLBr_01258|M.leprae_Br4923 ----------MNVPEPTLDDLPLRDNLRGKSPYGAMQLLVPVLLNTNENP
. : **
Mb3800|M.bovis_AF2122/97 FGP----LPSVRAAIDRATDTVNRYPDNGCVQLKAALARHLG----PDFA
Rv3772|M.tuberculosis_H37Rv FGP----LPSVRAAIDRATDTVNRYPDNGCVQLKAALARHLG----PDFA
MMAR_5326|M.marinum_M FGP----LPSVRAAIEHATQSINRYPDNGCLAVKAALARHVSSLSAADFG
MUL_4407|M.ulcerans_Agy99 FGP----LPSVRAAIEHATQSINRYPDNGCLAVKAALARHVSSLSAADFG
Mflv_1195|M.gilvum_PYR-GCK HGP----LPSVRAAIADAVGGINRYPDNGYVDLNERLAKYVG------FA
MSMEG_1491|M.smegmatis_MC2_155 YPP----LPAVKAALSAAVDAANRYPEFLPERLRSLIAGRVG------VD
Mvan_1355|M.vanbaalenii_PYR-1 FPP----LPAVRTALAASLDCLNRYPEFLPHRLRSLIAGRLG------AD
MAV_4421|M.avium_104 FPP----LPAVRAALIRSVCTANRYPEFLPERLRGVIAEHIG------VP
TH_3677|M.thermoresistible__bu FPP----LPAVRVALIEAVSAANRYPEFLPERLRSLIAERIG------VP
MAB_2168|M.abscessus_ATCC_1997 YRP----LPSVRDALRHDLAEVNRYPEFLPVVLPRLIADRVG------MT
MLBr_01258|M.leprae_Br4923 HPPTKALVDDVVRSVQKVAVDLHRYPDRDAVALRQDLASYLTAQTGIRLG
. * : * :: :***: : :* :
Mb3800|M.bovis_AF2122/97 PEHVAVGCGSVSLCQQLVQVTASVGDEVVFGWRSFELYPPQVRVAGAIPI
Rv3772|M.tuberculosis_H37Rv PEHVAVGCGSVSLCQQLVQVTASVGDEVVFGWRSFELYPPQVRVAGAIPI
MMAR_5326|M.marinum_M PEHIAVGCGSVSLCQQLVQITASVGDEVIFGWRSFELYPPQVQVAGATAI
MUL_4407|M.ulcerans_Agy99 PEHIAVGCGSVSLCQQLVQITASVGDEVIFGWRSFELYPPQVQVAGATAI
Mflv_1195|M.gilvum_PYR-GCK PENISVGCGSVSLCQQLIQITATVGDEVLFGWRSFEIYPLQVRTAGATPV
MSMEG_1491|M.smegmatis_MC2_155 EGQVIIGVGATGVIMQVLHAVTSPGDTMVMSAPTFDGYPIFAQMARLRSV
Mvan_1355|M.vanbaalenii_PYR-1 PEQVVIGAGATGVLMQVLGAVTSPGDTIVMSSPTFDGYPILADMARLRPV
MAV_4421|M.avium_104 AELVVLGAGATGVMLQVLQALTAPGDAIAMATPTFDGYPIVARMARLNTI
TH_3677|M.thermoresistible__bu PDQVILGGGATGVMLQVLHAIADEHDRIVMATPTFEGYPIVARMRRLEPV
MAB_2168|M.abscessus_ATCC_1997 PEEVVVGVGATGVALQVLQALCKPGDSLVFAHPTFDGYPILADIAGLIQV
MLBr_01258|M.leprae_Br4923 VENIWAANGSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIIADGTHTEWL
: . *: : *:: ::. :* . :
Mb3800|M.bovis_AF2122/97 QVPLTDHTFDLYAMLAAVTDRTR--LIFVCNPNNPTSTVVGPDALARFVE
Rv3772|M.tuberculosis_H37Rv QVPLTDHTFDLYAMLATVTDRTR--LIFVCNPNNPTSTVVGPDALARFVE
MMAR_5326|M.marinum_M QVPLTNHTFDLEAMLAAVTERTR--LIIVCNPNNPTSTVVQPEALAEFVR
MUL_4407|M.ulcerans_Agy99 QVPLTNHTFDLEAMLAAVTERTR--LIIVCDPNNPTSTVVQPEALAEFVR
Mflv_1195|M.gilvum_PYR-GCK QVPLTDHTFDLDAMLAAITDRTR--LIFVCNPNNPSSTVVDPGALARFVA
MSMEG_1491|M.smegmatis_MC2_155 TVDLDVYGHHNLHAMADAARDAR--VVVVCRPHNPTGTIEPAEDIERFLL
Mvan_1355|M.vanbaalenii_PYR-1 AVPLDAHGHHDLDAMADAAAQAR--VVVVCRPHNPTGTVESVADVERFLT
MAV_4421|M.avium_104 AVALDEHGQHDLDGLADAAARAR--VVVLCRPHNPTGTVESAAAVFRFLR
TH_3677|M.thermoresistible__bu EVPLAPDGRHDLTAMAEAAEHAR--VVALCRPHNPTGTVEPAAAIEGFLS
MAB_2168|M.abscessus_ATCC_1997 PIPLADNGMTDLDELLGAAYDADDAALVLCRPHNPTGTLIPADQVYEFVR
MLBr_01258|M.leprae_Br4923 ETVRADDFSLDVEAAVTAVADRKPDVVFIASPNNPSGQSISLADLRRLLD
. : :. *:**:. : ::
Mb3800|M.bovis_AF2122/97 A-VPAHILIAIDEAYVEYIRDGMRPDSLGLVRAHN-NVVVLRTFSKAYGL
Rv3772|M.tuberculosis_H37Rv A-VPAHILIAIDEAYVEYIRDGMRPDSLGLVRAHN-NVVVLRTFSKAYGL
MMAR_5326|M.marinum_M S-VPPHILVAIDEAYVEYLRDGTVPDSPHLVRTHS-NVVVLRTFSKAYGL
MUL_4407|M.ulcerans_Agy99 S-VPPHILVAIDEAYVEYLRDGTVPDSPHLVRTHS-NVVVLRTFSKAYGL
Mflv_1195|M.gilvum_PYR-GCK A-VPPHILVVLDEAYIEYVRGDLVPDSFGLVRAHS-NVVVLRTFSKAYGL
MSMEG_1491|M.smegmatis_MC2_155 ESIPEDTVVLLDEAYAEFLGPANRIDGPRLVERFR-NVVVVRTFSKAYGL
Mvan_1355|M.vanbaalenii_PYR-1 R-VGRDTVVLLDEAYVEFLAPEYRIDANALVRRFG-NVVVVRTFSKAYGL
MAV_4421|M.avium_104 R-VPRDTVVLLDEAYVEFTAPEHRIDVAALVACFP-NVVVVRTFSKAYGL
TH_3677|M.thermoresistible__bu R-LSADTVVLIDEAYVEFVAPGHRLDAVGLIRRFP-NVVGVRTFSKAYGL
MAB_2168|M.abscessus_ATCC_1997 Q-APPTSVVVLDEAYIEFVDPDEHLDIPGLLRLHP-NLVVLRTFSKAYGL
MLBr_01258|M.leprae_Br4923 V---VPGILIVDEAYGEFSS---RPSAVALVGEYPTKIVVTRTTSKAFAF
:: :**** *: . *: . ::* ** ***:.:
Mb3800|M.bovis_AF2122/97 AGLRIGYAIGHPDVITALDKVYVPFTVSSIGQAAAIASLDAADELLARTD
Rv3772|M.tuberculosis_H37Rv AGLRIGYAIGHPDVITALDKVYVPFTVSSIGQAAAIASLDAADELLARTD
MMAR_5326|M.marinum_M AGLRVGYAVGQPDVIAALDKVYVPFTVSSLAQAAAIASVQAADELLARTD
MUL_4407|M.ulcerans_Agy99 AGLRVGYAVGQPDVIAPLDKVYVPFTVSSLAQAAAIASVQAADELLARTD
Mflv_1195|M.gilvum_PYR-GCK AGLRIGYAVADPDIVTALSKVYVPFTATTVSQAAAIACLDAADELLERTD
MSMEG_1491|M.smegmatis_MC2_155 AGLRIGYGFCAPQLARQLWTMQLPFGIGLPALVAVAASYDAEGQLRQRIR
Mvan_1355|M.vanbaalenii_PYR-1 AGLRVGYGFCAPALGRTLWTMQLPFGTSLAALVAVAASFDADSQLRQRVR
MAV_4421|M.avium_104 AGLRIGYGVASPRLARPLWDQQLPFGIAITSLLAVTASYRAEDELSRRIR
TH_3677|M.thermoresistible__bu AGLRIGYAFGSPQLAARLWAMQLPFGMSITSLAAVAASYGADRQLRRRIR
MAB_2168|M.abscessus_ATCC_1997 AGLRIGYGLAAPAVCGVIRRYQLPFGMNRAAAVAVAASYAAEGELRVRVD
MLBr_01258|M.leprae_Br4923 AGGRLGYLIATPALVEAMLLVRLPYHLSSVTQAAARAALRHADDTLGSVA
** *:** . * : : :*: *. *. :
Mb3800|M.bovis_AF2122/97 TVVAERARVSAELRAAGFTLPPSQANFVWLPLGSRTQDFVEQAADARIVV
Rv3772|M.tuberculosis_H37Rv TVVAERARVSAELRAAGFTLPPSQANFVWLPLGSRTQDFVEQAADARIVV
MMAR_5326|M.marinum_M AVVAERGRVSAELRAAGFTVPPSQANFVWLPLEDRTTDFVTQAAKAHIVV
MUL_4407|M.ulcerans_Agy99 AVVAERGRVSAELRAAGFTVPPSQANFVWLPLEDRTTDFVTQAAKAHIVV
Mflv_1195|M.gilvum_PYR-GCK AVVAERTRVSAALREAGYDLPPSQANFVWLPLVGRAQQFAADAADARIIV
MSMEG_1491|M.smegmatis_MC2_155 MISAERRFLQMRLRSMGVYSTDGQANFVYLPAT--GVPWHDVFDDAGLKV
Mvan_1355|M.vanbaalenii_PYR-1 SISAERRRLQMRLRALGAYTTDAHANFVYLPSG--VTSWPEVFDGTGLQV
MAV_4421|M.avium_104 SITAERRRLRMRLSALGVYTTDAHANFVYLPPDPQGRAWQRVFARGGVRA
TH_3677|M.thermoresistible__bu MITTERRYLQTQLRALGVASTDAHANFVYLPAD--GRCWPEVFAGTDLRV
MAB_2168|M.abscessus_ATCC_1997 SIVYERDRLRARLAQMGAHIPESHANFVYLPARGDGERWMSILGTGDVVA
MLBr_01258|M.leprae_Br4923 ALIAERERVTKSLVHMGFRVIPSDANFVLFGHFSDAAGAWQHYLDTGVLI
: ** : * * ..**** : :
Mb3800|M.bovis_AF2122/97 RPYGTDG-VRVTVAAPEENDAFLRFARRWRSDQ------------
Rv3772|M.tuberculosis_H37Rv RPYGTDG-VRVTVAAPEENDAFLRFARRWRSDQ------------
MMAR_5326|M.marinum_M RPYGADG-VRVTIAAPEENDALLRFARCWITHRDGAR--------
MUL_4407|M.ulcerans_Agy99 RPYGADG-VRVTIAAPEENDALLRFARCWITHRDGAR--------
Mflv_1195|M.gilvum_PYR-GCK RPYGEDG-VRVTVASPDENDMFLDFARDWIARP------------
MSMEG_1491|M.smegmatis_MC2_155 RHYADGG-VRVTVGTRESTREVLR------ATRLALR--------
Mvan_1355|M.vanbaalenii_PYR-1 RHYAGGG-VRITVGSRASTRAVVH------AVAAAMG--------
MAV_4421|M.avium_104 RYYPDGA-ARLTVGSRASTVAVLS------AVGKATLERQVR---
TH_3677|M.thermoresistible__bu KHCPDGG-VRITVGDRSSTGAVLA------AIRGQR---------
MAB_2168|M.abscessus_ATCC_1997 KEYSDGG-VRLTVGDPREMAIVARRLLEVDEIGESAVSRMSRPAS
MLBr_01258|M.leprae_Br4923 RDVGIPGYLRATTGLAEENDAFLKASSEIAATELAPATTLGAS--
: . * * . . .