For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSIRLRAEMADLPAYAPGKTVPGAIKIASNETVHGPLPSVREAILKATDLINRYPDNGYLDLRERLAKHV NFAPENISVGCGSVSLCQQLIQITSSVGDEVLFAWRSFEIYPLQVRTAGATPVAVALRDHTHDLDAMLAA ITDRTRLIFVCNPNNPTSTVVDPGELARFVAAVPPHILVVLDEAYVEYIRDGLLPDSLGLVREHRNVVVL RTFSKAYGLAGLRVGYAVADPEIVTALGKVYVPFSATSVSQAAAIACLDAADELLARTDAVVAERTRVSD ALRAAGYTLPPSQANFVWLPLAERTLDFVARAADNRIIVRPYGEDGVRVTIGAPHENDAFLDFAQRWIAP GGAGPRTGDSA
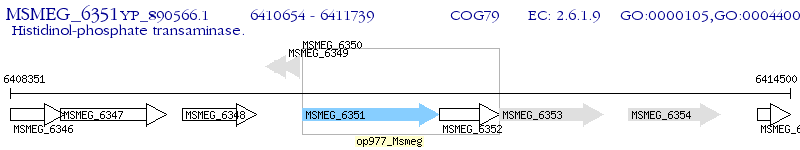
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6351 | hisC | - | 100% (361) | putative aminotransferase |
| M. smegmatis MC2 155 | MSMEG_1491 | - | 8e-44 | 35.81% (310) | histidinol-phosphate aminotransferase 2 |
| M. smegmatis MC2 155 | MSMEG_3206 | hisC | 1e-37 | 34.59% (344) | histidinol-phosphate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3800 | hisC2 | 1e-151 | 74.79% (349) | putative aminotransferase |
| M. gilvum PYR-GCK | Mflv_1195 | - | 1e-166 | 83.95% (349) | putative aminotransferase |
| M. tuberculosis H37Rv | Rv3772 | hisC2 | 1e-151 | 74.50% (349) | putative aminotransferase |
| M. leprae Br4923 | MLBr_01258 | hisC | 7e-38 | 31.98% (344) | histidinol-phosphate aminotransferase |
| M. abscessus ATCC 19977 | MAB_0220c | - | 1e-144 | 72.05% (347) | putative aminotransferase |
| M. marinum M | MMAR_5326 | hisC2 | 1e-142 | 71.19% (354) | histidinol-phosphate aminotransferase HisC2 |
| M. avium 104 | MAV_0250 | hisC | 1e-134 | 68.38% (351) | putative aminotransferase |
| M. thermoresistible (build 8) | TH_1662 | hisC2 | 1e-161 | 79.66% (349) | PROBABLE HISTIDINOL-PHOSPHATE AMINOTRANSFERASE HISC2 |
| M. ulcerans Agy99 | MUL_4407 | hisC2 | 1e-140 | 70.34% (354) | putative aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5611 | - | 1e-169 | 85.88% (347) | putative aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3800|M.bovis_AF2122/97 ---------MTARLRPELAGLPVYVPGKTVPGAIKLASNETVFGP----L
Rv3772|M.tuberculosis_H37Rv ---------MTARLRPELAGLPVYVPGKTVPGAIKLASNETVFGP----L
MMAR_5326|M.marinum_M ---------MTARLRPVLAGLPVYVPGKTVPGAIKLASNETVFGP----L
MUL_4407|M.ulcerans_Agy99 ---------MTAHLRPVLAGLPVYVPGKTVPGAIKLASNETVFGP----L
MAV_0250|M.avium_104 ---------MTARLRPELAGLPVYVPGKNVPGSIKLASNETVYGP----L
Mflv_1195|M.gilvum_PYR-GCK MRAGHKLSQVSARLRPELADLPAYTPGRTVPGAIKIASNETVHGP----L
Mvan_5611|M.vanbaalenii_PYR-1 ---------MSARLRPELADLPAYAPGKTVPGAIKIASNETVHGP----L
MSMEG_6351|M.smegmatis_MC2_155 ---------MSIRLRAEMADLPAYAPGKTVPGAIKIASNETVHGP----L
TH_1662|M.thermoresistible__bu ---------VTARLRPELADIPAYVPGKMVPGAIKIASNETVHPP----L
MAB_0220c|M.abscessus_ATCC_199 ---------MPARLRPELTELPAYTPGRNVPGAIKLASNETVQEP----L
MLBr_01258|M.leprae_Br4923 -MNVPEPTLDDLPLRDNLRGKSPYG-AMQLLVPVLLNTNENPHPPTKALV
** : . * . : .: : :**. * :
Mb3800|M.bovis_AF2122/97 PSVRAAIDRATDTVNRYPDNGCVQLKAALARHLG----PDFAPEHVAVGC
Rv3772|M.tuberculosis_H37Rv PSVRAAIDRATDTVNRYPDNGCVQLKAALARHLG----PDFAPEHVAVGC
MMAR_5326|M.marinum_M PSVRAAIEHATQSINRYPDNGCLAVKAALARHVSSLSAADFGPEHIAVGC
MUL_4407|M.ulcerans_Agy99 PSVRAAIEHATQSINRYPDNGCLAVKAALARHVSSLSAADFGPEHIAVGC
MAV_0250|M.avium_104 PSVHAAIERAVAIVNRYPDNACVDLKAALAMHLG----SDVAPEQIAVGS
Mflv_1195|M.gilvum_PYR-GCK PSVRAAIADAVGGINRYPDNGYVDLNERLAKYVG------FAPENISVGC
Mvan_5611|M.vanbaalenii_PYR-1 PSVRAAIEKAIDGINRYPDNGYVELRERLAKHVN------FAPEYISVGC
MSMEG_6351|M.smegmatis_MC2_155 PSVREAILKATDLINRYPDNGYLDLRERLAKHVN------FAPENISVGC
TH_1662|M.thermoresistible__bu PSVREAIARAGETLNRYPDNGYLDLRDRLARHVG------FAPEHISVGC
MAB_0220c|M.abscessus_ATCC_199 PSVRAALAEAGSLINRYPDNGYAELRSHLAKHVD------MPPEHIAVGC
MLBr_01258|M.leprae_Br4923 DDVVRSVQKVAVDLHRYPDRDAVALRQDLASYLTAQTGIRLGVENIWAAN
.* :: . ::****. :. ** :: . * : ..
Mb3800|M.bovis_AF2122/97 GSVSLCQQLVQVTASVGDEVVFGWRSFELYPPQVR-VAGAIPIQVPLTDH
Rv3772|M.tuberculosis_H37Rv GSVSLCQQLVQVTASVGDEVVFGWRSFELYPPQVR-VAGAIPIQVPLTDH
MMAR_5326|M.marinum_M GSVSLCQQLVQITASVGDEVIFGWRSFELYPPQVQ-VAGATAIQVPLTNH
MUL_4407|M.ulcerans_Agy99 GSVSLCQQLVQITASVGDEVIFGWRSFELYPPQVQ-VAGATAIQVPLTNH
MAV_0250|M.avium_104 GSVTLCQQLVQITSAAGDEVMMGWRSFECYLPIVQ-VAGAIAVKVPLREH
Mflv_1195|M.gilvum_PYR-GCK GSVSLCQQLIQITATVGDEVLFGWRSFEIYPLQVR-TAGATPVQVPLTDH
Mvan_5611|M.vanbaalenii_PYR-1 GSVSLCQQLIQITSTVGDEVLFGWRSFEIYPLQVR-TAGATPVQVPLTDH
MSMEG_6351|M.smegmatis_MC2_155 GSVSLCQQLIQITSSVGDEVLFAWRSFEIYPLQVR-TAGATPVAVALRDH
TH_1662|M.thermoresistible__bu GSVSLCQQLVQITAGAGDEVLFGWRSFEIYPLQVR-TAGAVPVQVPLTDH
MAB_0220c|M.abscessus_ATCC_199 GSVSLCQQLVQITATVGDEVLFGWRSFETYPLVVR-VAGATPVQVPLVDH
MLBr_01258|M.leprae_Br4923 GSNEILQQLLQAFGGPGRSAIGFVPSYSMHPIIADGTHTEWLETVRADDF
** : ***:* . * ..: *:. : . . * :.
Mb3800|M.bovis_AF2122/97 TFDLYAMLAAVTDRTR-LIFVCNPNNPTSTVVGPDALARFVEAVPAHILI
Rv3772|M.tuberculosis_H37Rv TFDLYAMLATVTDRTR-LIFVCNPNNPTSTVVGPDALARFVEAVPAHILI
MMAR_5326|M.marinum_M TFDLEAMLAAVTERTR-LIIVCNPNNPTSTVVQPEALAEFVRSVPPHILV
MUL_4407|M.ulcerans_Agy99 TFDLEAMLAAVTERTR-LIIVCDPNNPTSTVVQPEALAEFVRSVPPHILV
MAV_0250|M.avium_104 TYDLDAMLAAITDRTR-LIFVCNPNNPTSTVVDPDALVRFVDAVPADILI
Mflv_1195|M.gilvum_PYR-GCK TFDLDAMLAAITDRTR-LIFVCNPNNPSSTVVDPGALARFVAAVPPHILV
Mvan_5611|M.vanbaalenii_PYR-1 TFDLDAMLAAITDRTR-LIFVCNPNNPTSTVVDPDALARFVEAVPPHIMV
MSMEG_6351|M.smegmatis_MC2_155 THDLDAMLAAITDRTR-LIFVCNPNNPTSTVVDPGELARFVAAVPPHILV
TH_1662|M.thermoresistible__bu TFDLDAILAAITDRTR-LIFICNPNNPTSTVVDPDALARFVAAVPEHILI
MAB_0220c|M.abscessus_ATCC_199 TYDLAAMAAAVTDVTR-LIFVCNPNNPTGTVVRPAELRRFVESVPPHILI
MLBr_01258|M.leprae_Br4923 SLDVEAAVTAVADRKPDVVFIASPNNPSGQSISLADLRRLLDVVPG--IL
: *: * ::::: . ::::..****:. : * .:: ** ::
Mb3800|M.bovis_AF2122/97 AIDEAYVEYIRDGMRPDSLGLVRAH-NNVVVLRTFSKAYGLAGLRIGYAI
Rv3772|M.tuberculosis_H37Rv AIDEAYVEYIRDGMRPDSLGLVRAH-NNVVVLRTFSKAYGLAGLRIGYAI
MMAR_5326|M.marinum_M AIDEAYVEYLRDGTVPDSPHLVRTH-SNVVVLRTFSKAYGLAGLRVGYAV
MUL_4407|M.ulcerans_Agy99 AIDEAYVEYLRDGTVPDSPHLVRTH-SNVVVLRTFSKAYGLAGLRVGYAV
MAV_0250|M.avium_104 AIDEAYVEYIRDGLLPNSLELALSR-SNVVVLRTFSKAYGLAGLRVGYAI
Mflv_1195|M.gilvum_PYR-GCK VLDEAYIEYVRGDLVPDSFGLVRAH-SNVVVLRTFSKAYGLAGLRIGYAV
Mvan_5611|M.vanbaalenii_PYR-1 VVDEAYVEYIRDGLAPDSFGLVRAH-SNVVVLRTFSKAYGLAGLRIGYAV
MSMEG_6351|M.smegmatis_MC2_155 VLDEAYVEYIRDGLLPDSLGLVREH-RNVVVLRTFSKAYGLAGLRVGYAV
TH_1662|M.thermoresistible__bu ALDEAYIEYVRPPFVPDSLGLVRAH-DNVVVLRTFSKAYGLAGLRVGYAV
MAB_0220c|M.abscessus_ATCC_199 AIDEAYVEYVREDFT-DSLALVREH-PNVVVLRTFSKAYGLAGLRVGYAV
MLBr_01258|M.leprae_Br4923 IVDEAYGEFS---SRPSAVALVGEYPTKIVVTRTTSKAFAFAGGRLGYLI
:**** *: .: *. ::** ** ***:.:** *:** :
Mb3800|M.bovis_AF2122/97 GHPDVITALDKVYVPFTVSSIGQAAAIASLDAADELLARTDTVVAERARV
Rv3772|M.tuberculosis_H37Rv GHPDVITALDKVYVPFTVSSIGQAAAIASLDAADELLARTDTVVAERARV
MMAR_5326|M.marinum_M GQPDVIAALDKVYVPFTVSSLAQAAAIASVQAADELLARTDAVVAERGRV
MUL_4407|M.ulcerans_Agy99 GQPDVIAPLDKVYVPFTVSSLAQAAAIASVQAADELLARTDAVVAERGRV
MAV_0250|M.avium_104 GHPELITALDKVVMPFAVTNVAQAAAIASLEASGELMARTDALVAERTRV
Mflv_1195|M.gilvum_PYR-GCK ADPDIVTALSKVYVPFTATTVSQAAAIACLDAADELLERTDAVVAERTRV
Mvan_5611|M.vanbaalenii_PYR-1 ADPDIIAALSKVYVPFTATSVSQAAAIACLDAADELLERTDAVVAERTRV
MSMEG_6351|M.smegmatis_MC2_155 ADPEIVTALGKVYVPFSATSVSQAAAIACLDAADELLARTDAVVAERTRV
TH_1662|M.thermoresistible__bu AAPEIVTALGKVYVPFSVSTLAQAAAIASLDAADELLARTDEVVAERARV
MAB_0220c|M.abscessus_ATCC_199 GDPDVITTLGKVYVPFSASSLAQAAAVASLGAAEELLARTNDVVTERARV
MLBr_01258|M.leprae_Br4923 ATPALVEAMLLVRLPYHLSSVTQAAARAALRHADDTLGSVAALIAERERV
. * :: .: * :*: :.: **** *.: : : : . :::** **
Mb3800|M.bovis_AF2122/97 SAELRAAGFTLPPSQANFVWLPLGSRTQDFVEQAADARIVVRPYGTDG-V
Rv3772|M.tuberculosis_H37Rv SAELRAAGFTLPPSQANFVWLPLGSRTQDFVEQAADARIVVRPYGTDG-V
MMAR_5326|M.marinum_M SAELRAAGFTVPPSQANFVWLPLEDRTTDFVTQAAKAHIVVRPYGADG-V
MUL_4407|M.ulcerans_Agy99 SAELRAAGFTVPPSQANFVWLPLEDRTTDFVTQAAKAHIVVRPYGADG-V
MAV_0250|M.avium_104 STTLRDAGFELPPSQANFLWLPLGSRTEDFVQEAANARLVVRPFASEG-V
Mflv_1195|M.gilvum_PYR-GCK SAALREAGYDLPPSQANFVWLPLVGRAQQFAADAADARIIVRPYGEDG-V
Mvan_5611|M.vanbaalenii_PYR-1 SAALREAGYDLPPSQANFVWLPLVGRAQQFAADAANSRVIVRPYGEDG-V
MSMEG_6351|M.smegmatis_MC2_155 SDALRAAGYTLPPSQANFVWLPLAERTLDFVARAADNRIIVRPYGEDG-V
TH_1662|M.thermoresistible__bu TAALRDAGYTVPESGANFVWLPLAERTVDFVNSAAEDRLLVRPYGTDG-V
MAB_0220c|M.abscessus_ATCC_199 TSALREAGYQVPPSQANFVWLPLGERSTEFAQASAEARIIVRPFGTDG-V
MLBr_01258|M.leprae_Br4923 TKSLVHMGFRVIPSDANFVLFGHFSDAAGAWQHYLDTGVLIRDVGIPGYL
: * *: : * ***: : : . :::* . * :
Mb3800|M.bovis_AF2122/97 RVTVAAPEENDAFLRFARRWRSDQ----------
Rv3772|M.tuberculosis_H37Rv RVTVAAPEENDAFLRFARRWRSDQ----------
MMAR_5326|M.marinum_M RVTIAAPEENDALLRFARCWITHRDGAR------
MUL_4407|M.ulcerans_Agy99 RVTIAAPEENDALLRFARCWITHRDGAR------
MAV_0250|M.avium_104 RVTIGAPAENDALLQFACDWIARTER--------
Mflv_1195|M.gilvum_PYR-GCK RVTVASPDENDMFLDFARDWIARP----------
Mvan_5611|M.vanbaalenii_PYR-1 RVTIAAPHENDAFLGFARGWDGLR----------
MSMEG_6351|M.smegmatis_MC2_155 RVTIGAPHENDAFLDFAQRWIAPGGAGPRTGDSA
TH_1662|M.thermoresistible__bu RVTVAAPHENDAFLSFATRWISER----------
MAB_0220c|M.abscessus_ATCC_199 RVTIGAPMENDAFLKFSRAWR-------------
MLBr_01258|M.leprae_Br4923 RATTGLAEENDAFLKASSEIAATELAPATTLGAS
*.* . . *** :* :