For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGTIWGTGRNFPVADGGPPDVPQLFVKNTHSVIGPGEPLQLTGSVATRVVAEGELAIVIGDQCRDLVTAE QARTVIAGYWVANDVTARDLQEVDDHWSRGKGLDGFCPLGVEMVAPRDMPTWKDVMIRTWVDDELVQDES VAAAYVGPEELVCWASQQFTLHPGDIFLLGTPAYRPAGDESRAVLKPGHTARVEITGIGELITPVI
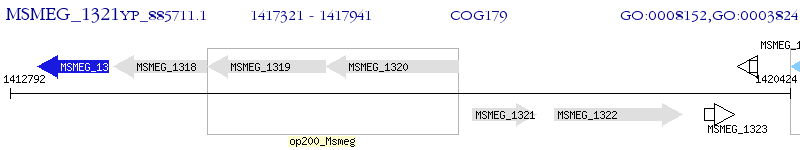
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1321 | - | - | 100% (206) | 5-carboxymethyl-2-hydroxymuconate delta-isomerase |
| M. smegmatis MC2 155 | MSMEG_2382 | - | 3e-29 | 40.84% (191) | 5-carboxymethyl-2-hydroxymuconate delta-isomerase |
| M. smegmatis MC2 155 | MSMEG_2007 | - | 1e-18 | 32.43% (185) | putative HpcE protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3017c | - | 5e-29 | 37.02% (235) | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase |
| M. gilvum PYR-GCK | Mflv_4230 | - | 2e-28 | 40.84% (191) | 5-carboxymethyl-2-hydroxymuconate Delta-isomerase |
| M. tuberculosis H37Rv | Rv2993c | - | 5e-29 | 37.02% (235) | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase |
| M. leprae Br4923 | MLBr_01689 | - | 2e-27 | 37.30% (185) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3299c | - | 1e-32 | 42.93% (191) | hypothetical protein MAB_3299c |
| M. marinum M | MMAR_1718 | - | 1e-27 | 39.27% (191) | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase |
| M. avium 104 | MAV_3844 | - | 6e-29 | 39.38% (193) | 5-carboxymethyl-2-hydroxymuconate delta-isomerase |
| M. thermoresistible (build 8) | TH_2040 | - | 4e-29 | 42.70% (185) | POSSIBLE 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE (HHDD |
| M. ulcerans Agy99 | MUL_1956 | - | 1e-26 | 38.22% (191) | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase |
| M. vanbaalenii PYR-1 | Mvan_2132 | - | 8e-28 | 39.27% (191) | 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3017c|M.bovis_AF2122/97 --------------------------MTAREIAEHPFGTPTFTGRSWPLA
Rv2993c|M.tuberculosis_H37Rv --------------------------MTAREIAEHPFGTPTFTGRSWPLA
MMAR_1718|M.marinum_M MRLGRIASPDGVAFASIEGELDNPGAMTAREIAEHPFGTPNFTGRSWPLA
MUL_1956|M.ulcerans_Agy99 --------------------------MTAREIAEDPFGTPNFTGRSWPLA
MAV_3844|M.avium_104 MRLGRIASPDGVAFVSIEGELDNPAEMIAREIAEHPFGTPNFTGRSWPLA
MLBr_01689|M.leprae_Br4923 --------------------------MFARELAEYPFGELTFTGRSWPLA
Mflv_4230|M.gilvum_PYR-GCK MRLGRIASPDGVAFVAVEGPPDDPAAAVAREIAEHPFGTPTFTGRQWPLA
Mvan_2132|M.vanbaalenii_PYR-1 MRLGRIASPDGVAFVAIEGPPDDPGGAVAREIAEHPFGNPTFTGRQWPLA
TH_2040|M.thermoresistible__bu MRLGRIASPDGVAFVSIEGPPDEP-TAVAREIAEHPFGTPEFTGRQWSLA
MAB_3299c|M.abscessus_ATCC_199 MRLGRIASPDGVAFVSIEGEDGREESAIAREIAEHPFGTPTFTGRQWPLA
MSMEG_1321|M.smegmatis_MC2_155 ------------------------------------MGTIWGTGRNFPVA
:* ***.:.:*
Mb3017c|M.bovis_AF2122/97 DVRLLAPILASKVVCVGKNYADHIAEMGGRP---PADPVIFLKPNTAIIG
Rv2993c|M.tuberculosis_H37Rv DVRLLAPILASKVVCVGKNYADHIAEMGGRP---PADPVIFLKPNTAIIG
MMAR_1718|M.marinum_M DVRLLAPILASKVVCVGKNYTDHIAEMGGQTGPTPADPVIFLKPNTAIVG
MUL_1956|M.ulcerans_Agy99 DVRLLAPILASKVVCVGKNYTGHIAEMGGQTGPTPADPVIFLKSNTAIVG
MAV_3844|M.avium_104 DVRLLAPILASKVVCVGKNYADHIAEMGEATGPVPADPVIFLKPNTAIIG
MLBr_01689|M.leprae_Br4923 DVRLLAPMLASKVVCIGKNYADHIVEMGGQTGLASADPVIFLKPNTAIIG
Mflv_4230|M.gilvum_PYR-GCK DVRLLAPILASKVVCMGKNYAAHIEEMGGGT---FEDPIIFLKPNTAIIG
Mvan_2132|M.vanbaalenii_PYR-1 DVRLLAPILASKVVCMGKNYAAHIEEMGGGT---FEDPIIFLKPNTAIIG
TH_2040|M.thermoresistible__bu DVRLLAPILASKVVCMGKNYAAHVDEMGGET---VPHPVIFIKPNTSIIG
MAB_3299c|M.abscessus_ATCC_199 DVRLLAPILASKVVAMGKNYAAHAAEMGGDP---PESPVIFIKPNTSIIG
MSMEG_1321|M.smegmatis_MC2_155 D--------------------------GGPP----DVPQLFVKNTHSVIG
* * . * :*:* . :::*
Mb3017c|M.bovis_AF2122/97 PNTPIRLP-ANASPVHFEGELAIVIGRACKD-VPAAQAVDNILGYTIGND
Rv2993c|M.tuberculosis_H37Rv PNTPIRLP-ANASPVHFEGELAIVIGRACKD-VPAAQAVDNILGYTIGND
MMAR_1718|M.marinum_M PNVPIRLP-ANASPVHFEGELAIVIGRPCKD-VSAAQAVDNILGYTIGND
MUL_1956|M.ulcerans_Agy99 PNVPIRLP-ANASPVHFEGELAIVIGRPCKD-VSAAQAVDNILGYTIGND
MAV_3844|M.avium_104 PNVPIRLP-ANASPVHFEGELAVVIGRPCKD-VSAAQAAENILGYTIGND
MLBr_01689|M.leprae_Br4923 PNVPIRLP-ANASPVHFEGELALVVGRSCKD-VPAVQAVDCILGYTIGND
Mflv_4230|M.gilvum_PYR-GCK PYTPIQLP-ADADPVHFEGELAAVIGRPCKD-VPASRAAENILGYTIAND
Mvan_2132|M.vanbaalenii_PYR-1 PNTPIQLP-ADANPVHFEGELAAVIGRPCKD-VPASRAAENILGYTIAND
TH_2040|M.thermoresistible__bu PNVPIQLP-VDADPVHHEGELAVVIGRPCKD-VPAARAADVILGYTVAND
MAB_3299c|M.abscessus_ATCC_199 PGLPIQLP-PSATEVHHEGELAIVIGRPCKD-VPAARAAENILGYTIGND
MSMEG_1321|M.smegmatis_MC2_155 PGEPLQLTGSVATRVVAEGELAIVIGDQCRDLVTAEQARTVIAGYWVAND
* *::*. * * ***** *:* *:* *.* :* * ** :.**
Mb3017c|M.bovis_AF2122/97 VSARDQQQSDGQWTRAKGHDTFCPVGPWIVTDLAPFDPADLELRTVVNGD
Rv2993c|M.tuberculosis_H37Rv VSARDQQQSDGQWTRAKGHDTFCPVGPWIVTDLAPFDPADLELRTVVNGD
MMAR_1718|M.marinum_M VSARDQQKSDGQWTRAKGHDTFCPVGPWIVTDV---DPADLELRTEVNGA
MUL_1956|M.ulcerans_Agy99 VSARDQQKSDGQWTRAKGHDTFCPVGPWIVTDV---DPAGLELRTEVNGA
MAV_3844|M.avium_104 VSARDQQRSDGQWTRAKGHDTFCPVGPWIVTDLNPLDPGDLELRTEVNGE
MLBr_01689|M.leprae_Br4923 MSARDQQRADGQWTRAKGHDTFCPVGPWIVTDLGPLDPADLELRTAVNGE
Mflv_4230|M.gilvum_PYR-GCK VSARDQQKKDGQWMRAKGHDTFCPVGPWIVTDI---DPSDLEIRTEVNGE
Mvan_2132|M.vanbaalenii_PYR-1 VSARDQQQKDGQWMRAKGHDTFCPVGPWIVTDL---DPSDLEIRTEVNGD
TH_2040|M.thermoresistible__bu VTARNQQRADGQWTRAKGHDTFCPVGPWIVTDV---DPTDLELRTEVNGE
MAB_3299c|M.abscessus_ATCC_199 VSARDHQRADGQWTRAKGHDTFCPLGPWIVTDL---DPADLEIRTEVNGQ
MSMEG_1321|M.smegmatis_MC2_155 VTARDLQEVDDHWSRGKGLDGFCPLGVEMVAPRDMPTWKDVMIRTWVDDE
::**: *. *.:* *.** * ***:* :*: .: :** *:.
Mb3017c|M.bovis_AF2122/97 VKQHARTSLMIHDIGAIVEWISAIMTLLPGDLILTGTPAG--VG-----P
Rv2993c|M.tuberculosis_H37Rv VKQHARTSLMIHDIGAIVEWISAIMTLLPGDLILTGTPAG--VG-----P
MMAR_1718|M.marinum_M VKQHARTSLMIHDVGAIVEWISAVMTLLPGDLILTGTPAG--VG-----P
MUL_1956|M.ulcerans_Agy99 VKQHARTSLMIHDVGAIVEWISAVMTLLPGDLILTGTPAG--VG-----P
MAV_3844|M.avium_104 VKQRSRTSLLIHDIGSIVEWISAVMTLLPGDLILTGTPAG--VG-----P
MLBr_01689|M.leprae_Br4923 VKQHSRTSLMIHGIGAIMEWISAVMTLLPGDLILTGTPAG--VG-----P
Mflv_4230|M.gilvum_PYR-GCK LKQHSRTSLMIHDIGAIIEWISSVMTLLPGDLILTGTPEG--VG-----P
Mvan_2132|M.vanbaalenii_PYR-1 VKQRSRTSLMIHDIGAIIEWISAVMTLLPGDLILTGTPEG--VG-----P
TH_2040|M.thermoresistible__bu VRQRSRTSLMIHDIGAIIEWVSRVMTLLPGDLILTGTPEG--VG-----P
MAB_3299c|M.abscessus_ATCC_199 VRQRSRTSLLLHDVGAIVEWVSAVMTLLPGDVILTGTPEG--VG-----P
MSMEG_1321|M.smegmatis_MC2_155 LVQDESVAAAYVGPEELVCWASQQFTLHPGDIFLLGTPAYRPAGDESRAV
: * .: . :: * * :** ***::* *** .*
Mb3017c|M.bovis_AF2122/97 IEDGDTVSITIEGIGTLTNPVVRKGKP
Rv2993c|M.tuberculosis_H37Rv IEDGDTVSITIEGIGTLTNPVVRKGKP
MMAR_1718|M.marinum_M IEDGDTVSITIEGIGTLTNPVVRKGKS
MUL_1956|M.ulcerans_Agy99 IEDGDTVSITIEGIDTLTNPVVRKGKS
MAV_3844|M.avium_104 IEDGDTVSITIEGIGTLTNPVVRKGKS
MLBr_01689|M.leprae_Br4923 VEDGDIVSITVEGIGTLTNPVVRKGKS
Mflv_4230|M.gilvum_PYR-GCK LEHGDTVSITVEGIGTLSNPVVRKGKS
Mvan_2132|M.vanbaalenii_PYR-1 LEHGDSVSITVEGIGTLTNPVVRKGRS
TH_2040|M.thermoresistible__bu IEHGDTVAVTIENIGTLTNPVVRKGKK
MAB_3299c|M.abscessus_ATCC_199 IVDGDTVSVTIEGIGTLSNPVVRKAK-
MSMEG_1321|M.smegmatis_MC2_155 LKPGHTARVEITGIGELITPVI-----
: *. . : : .*. * .**: