For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTLVAGRTFPWVRRSSAGGPVQVDTDTPKPQWRLAVRAFSRDRMAVVSLVVLVIVTLAAVFAPLLTPYS PTAGDPVDRLAGIGTEGHLLGLDGQGRDIWTRLLYGGRNSLMTAVVPVFVVFPLALMIGLFAGYRRSRAG EVLMRILDVLFAFPLVLLAIALSAVLGAGLGNVMLAIGITLIPYMARVAYTATVSEASKDYIEAARAYGA NPLLLMFRELMPNVVVQLLVYSTTLCGLMIVVASGLSFLGVGVIPPTPDWGIMTADGKNVLLEGIYHVAT IPGLLILVVSLAFNLVGDGIRDALDPRKQTN
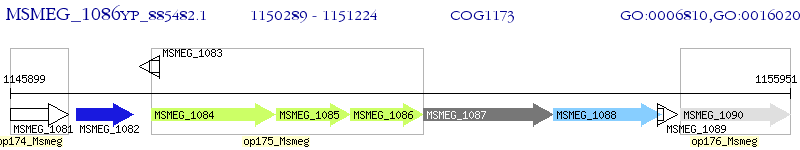
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1086 | - | - | 100% (311) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_6867 | - | 1e-55 | 44.15% (265) | oligopeptide ABC transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_4356 | - | 1e-46 | 37.28% (287) | inner membrane ABC transporter permease protein YddQ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 1e-31 | 31.03% (261) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 2e-47 | 37.41% (278) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 1e-31 | 31.03% (261) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 3e-27 | 29.56% (274) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0721 | - | 3e-32 | 30.91% (275) | oligopeptide ABC transporter, permease protein |
| M. marinum M | MMAR_5152 | dppC | 6e-32 | 30.65% (261) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_0466 | dppC | 6e-32 | 30.91% (275) | ABC transporter, permease protein DppC |
| M. thermoresistible (build 8) | TH_2705 | - | 7e-45 | 36.00% (275) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 2e-32 | 31.44% (264) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 7e-49 | 38.38% (284) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2759|M.gilvum_PYR-GCK -----------------MTTTDPQS------TRLSSWRLLALNPVTLISA
Mvan_3775|M.vanbaalenii_PYR-1 -----------------MTTTDPND------TRLSSWRLLAGNPVTLVSA
TH_2705|M.thermoresistible__bu -----------------MTADDLTGGEPDAVSRVAAWRLLAANPVTLLSA
Mb3688c|M.bovis_AF2122/97 ----------------------------------------------MIAA
Rv3664c|M.tuberculosis_H37Rv ----------------------------------------------MIAA
MAV_0466|M.avium_104 ---------------------MAERIRARGGFWRETWRRLRRRPKFIGAG
MMAR_5152|M.marinum_M -------------------------MAELGGFWFDILGGLRRRPKFVIAA
MUL_4238|M.ulcerans_Agy99 -------------------------MAELRGFWFDILGGLRRRPKFVIAA
MSMEG_1086|M.smegmatis_MC2_155 MSTLVAGRTFPWVRRSSAGGPVQVDTDTPKPQWRLAVRAFSRDRMAVVSL
MLBr_01123|M.leprae_Br4923 --------------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSL
MAB_0721|M.abscessus_ATCC_1997 -------------MTTSVSKPHGNRLRRLPGADLLRSVHGLQRWTLVVGL
.
Mflv_2759|M.gilvum_PYR-GCK LVLIAVAVVAVTANWIIPYGVNDVDVP----NALAPP-SGSHWFGTDELG
Mvan_3775|M.vanbaalenii_PYR-1 LVLVAVTVVAVTANWLAPYGVNDVDVP----NALAPP-SASHWFGTDELG
TH_2705|M.thermoresistible__bu ALLALLVAVAVTAPWLAPYGVNDINVP----EALQPP-SAAHWFGTDELG
Mb3688c|M.bovis_AF2122/97 ALILLILVVAAFPSLFTAADPTYADPS----QSMLAP-SAAHWFGTDLQG
Rv3664c|M.tuberculosis_H37Rv ALILLILVVAAFPSLFTAADPTYADPS----QSMLAP-SAAHWFGTDLQG
MAV_0466|M.avium_104 LLILVILAVALFPALFTAADPTYADPA----QSLLPP-SRTHWFGTDLQG
MMAR_5152|M.marinum_M VLIVFILVVAAFPSLFTGIDPSYADPS----QSMLSP-STAHWFGTDLQG
MUL_4238|M.ulcerans_Agy99 VLIVFILFVAAFPSLFTGIDPSYADPS----QSMLSP-STAHWFGTDLQG
MSMEG_1086|M.smegmatis_MC2_155 VVLVIVTLAAVFAPLLTPYSPTAGDPV----DRLAGIGTEGHLLGLDGQG
MLBr_01123|M.leprae_Br4923 MLLVLLFVACYLLPSLLPYSYDDLDFN----ALLQPP-NTRHWLGTNALG
MAB_0721|M.abscessus_ATCC_1997 VLVAGFIAIAVLAPVLAPYGFSQTSADGLDFVRQQAP-SWQHWFGTSVRG
:: . . : . . . * :* . *
Mflv_2759|M.gilvum_PYR-GCK RDVLSRVLVATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRV
Mvan_3775|M.vanbaalenii_PYR-1 RDVLSRVLVAVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRV
TH_2705|M.thermoresistible__bu RDVLSRVLVAIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRL
Mb3688c|M.bovis_AF2122/97 HDIYSRTVYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRV
Rv3664c|M.tuberculosis_H37Rv HDIYSRTVYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRV
MAV_0466|M.avium_104 HDVYARTVYGARASVTVGLGAAAIVFVVGGALGALAGFYGGWIDAVVSRV
MMAR_5152|M.marinum_M HDIYARTVYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRI
MUL_4238|M.ulcerans_Agy99 HDIYARTVYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRI
MSMEG_1086|M.smegmatis_MC2_155 RDIWTRLLYGGRNSLMTAVVPVFVVFPLALMIGLFAGYRRSRAGEVLMRI
MLBr_01123|M.leprae_Br4923 QDLLAQTLRGMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWV
MAB_0721|M.abscessus_ATCC_1997 EDVLSRVLFGARTALLVIVTSLVVSLPAGVVLGLTSGYLGGWLDRALVLV
.*: :: : . : :: . :* :*: . .: :
Mflv_2759|M.gilvum_PYR-GCK VDVMFAFP-VLLLALAVVAIL-----GPG-VTTTILAIGIVYTPIFARVA
Mvan_3775|M.vanbaalenii_PYR-1 VDVMFAFP-VLLLALAVVAIL-----GPG-VTTTILAIAIVYTPIFARVA
TH_2705|M.thermoresistible__bu VDVLFAFP-VLLLALAIVAIL-----GPG-MTTTMLAIGIVFTPIFARVA
Mb3688c|M.bovis_AF2122/97 TDVFLGLP-LLLAAIVLMQVM-----HHRTVWTVIAILALFGWPQVARIA
Rv3664c|M.tuberculosis_H37Rv TDVFLGLP-LLLAAIVLMQVM-----HHRTVWTVIAILALFGWPQVARIA
MAV_0466|M.avium_104 TDVFFGLP-LLLAAIVLMQVL-----HHRTVWTVIAILALFGWPQVARIA
MMAR_5152|M.marinum_M TDIFFGLP-LLLAAIVLMQVM-----HHRTVWTVIAILALFGWPQIARIA
MUL_4238|M.ulcerans_Agy99 TDIFFGLP-LLLAAIVLMQVM-----HHRTVWTVIAILALFGWPQIARIA
MSMEG_1086|M.smegmatis_MC2_155 LDVLFAFP-LVLLAIALSAVL-----GAG-LGNVMLAIGITLIPYMARVA
MLBr_01123|M.leprae_Br4923 VDLLLVVPSFVLIAIMSPQTK-----SSDNIMLLVLLLAGFGWMVSSRMV
MAB_0721|M.abscessus_ATCC_1997 MDALYAMPSLLLAIVVSIAVTGGQSSTGGGILAAAIAIMAVFVPQYFRVV
* : .* .:* : : : *:.
Mflv_2759|M.gilvum_PYR-GCK RASTLGVRTEPYVSVSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFA
Mvan_3775|M.vanbaalenii_PYR-1 RASTLGVRTEPYVSASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFA
TH_2705|M.thermoresistible__bu RAGTLGVRVEPFVQASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFA
Mb3688c|M.bovis_AF2122/97 RGAVLEVRASDYVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIF
Rv3664c|M.tuberculosis_H37Rv RGAVLEVRASDYVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIF
MAV_0466|M.avium_104 RGAVLAVRASDYVLAAKALGMSRFQILIRHALPNALGPVIAVATIALGLF
MMAR_5152|M.marinum_M RSSVLEVRGSDFVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIF
MUL_4238|M.ulcerans_Agy99 RSSVLEVRGSDFVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIF
MSMEG_1086|M.smegmatis_MC2_155 YTATVSEASKDYIEAARAYGANPLLLMFRELMPNVVVQLLVYSTTLCGLM
MLBr_01123|M.leprae_Br4923 RGMTMSLRKSEFIRAGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSA
MAB_0721|M.abscessus_ATCC_1997 RNATVGVKQEPFVDAARVTGAGTPRILFRHILSNVTSSLPVIITLNGAEA
.: . :: ..: * . :: . :.* : : .
Mflv_2759|M.gilvum_PYR-GCK ILSEAALSFLGLGIQP-PQPSLGRMIFDSQ-GFVTMAWWMAVFPGAAIFL
Mvan_3775|M.vanbaalenii_PYR-1 ILSEAALSFLGLGIQP-PQPSLGRMIFDSQ-GFVTMAWWMAVFPGAAIFV
TH_2705|M.thermoresistible__bu ILAEAALSFLGLGIQP-PEPSLGRMIFDSQ-GFVTLAWWMAVFPGAAIVA
Mb3688c|M.bovis_AF2122/97 IVTEATLSYLGVGLPT-SVVSWGGDINVAQTRLRSGS-PILFYPAGALAI
Rv3664c|M.tuberculosis_H37Rv IVTEATLSYLGVGLPT-SVVSWGGDINVAQTRLRSGS-PILFYPAGALAI
MAV_0466|M.avium_104 IVTEATLSYLGVGLPP-SVVSWGGDINLAQTRLRAGS-PILFYPAGALAA
MMAR_5152|M.marinum_M IVTEATLSYLGVGLPP-TVVSWGGDINVAQMRLRAGS-PILFYPAGALAV
MUL_4238|M.ulcerans_Agy99 IVTEATLSYLGVGLPP-TVVSWGDDINVAQMRLRAGS-PILFYPAGALAV
MSMEG_1086|M.smegmatis_MC2_155 IVVASGLSFLGVGVIP-PTPDWGIMTADGKNVLLEGIYHVATIPGLLILV
MLBr_01123|M.leprae_Br4923 ILAETGLSFLGFGIKP-PDVSLGTLIANGTQSATTFP-WVFLFPAGVLVV
MAB_0721|M.abscessus_ATCC_1997 ILTLAGLGFLGFGIEPTQAAEWGYDLNKALSDVANGIWWTGVFPGIAIVL
*: : *.:**.*: . . * . *. :
Mflv_2759|M.gilvum_PYR-GCK IVLAFNLFGDGLRDVLDPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 IVLAFNLLGDGLRDVLDPKQRTTVEARRRER-------------
TH_2705|M.thermoresistible__bu AVLACNLFGDGLRDVLDPKQRTVIETQRRARQRPGRGRRAGSAW
Mb3688c|M.bovis_AF2122/97 TVLAFMMMGDALRDALDPASRAWRA-------------------
Rv3664c|M.tuberculosis_H37Rv TVLAFMMMGDALRDALDPASRAWRA-------------------
MAV_0466|M.avium_104 TVLAFMMMGDALRDALDPASRAWRA-------------------
MMAR_5152|M.marinum_M TVLAFMMMGDALRDALDPASRAWRA-------------------
MUL_4238|M.ulcerans_Agy99 TVLAFMMMGDALRDALDPASRAWRA-------------------
MSMEG_1086|M.smegmatis_MC2_155 VSLAFNLVGDGIRDALDPRKQTN---------------------
MLBr_01123|M.leprae_Br4923 ILVCANLTGDGLRDAFDPVSKHLRR-------------------
MAB_0721|M.abscessus_ATCC_1997 VVLGMTLVGESLNEVINPLLRTRRGEA-----------------
: : *:.:.:.::* :