For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSVSKPHGNRLRRLPGADLLRSVHGLQRWTLVVGLVLVAGFIAIAVLAPVLAPYGFSQTSADGLDFVR QQAPSWQHWFGTSVRGEDVLSRVLFGARTALLVIVTSLVVSLPAGVVLGLTSGYLGGWLDRALVLVMDAL YAMPSLLLAIVVSIAVTGGQSSTGGGILAAAIAIMAVFVPQYFRVVRNATVGVKQEPFVDAARVTGAGTP RILFRHILSNVTSSLPVIITLNGAEAILTLAGLGFLGFGIEPTQAAEWGYDLNKALSDVANGIWWTGVFP GIAIVLVVLGMTLVGESLNEVINPLLRTRRGEA
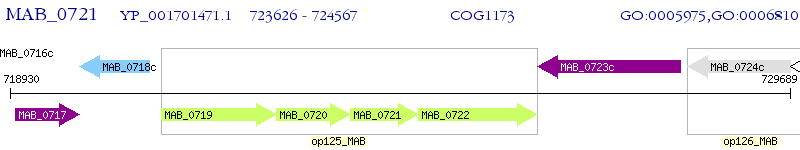
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0721 | - | - | 100% (313) | oligopeptide ABC transporter, permease protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 6e-29 | 30.29% (274) | peptide ABC transporter DppC |
| M. abscessus ATCC 19977 | MAB_0427 | - | 2e-05 | 24.43% (176) | peptide ABC transporter transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1313c | oppC | 3e-35 | 32.18% (289) | oligopeptide-transport integral membrane protein ABC |
| M. gilvum PYR-GCK | Mflv_2759 | - | 4e-48 | 40.00% (280) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1282c | oppC | 3e-35 | 32.18% (289) | oligopeptide-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01123 | - | 1e-33 | 30.36% (280) | putative binding-protein dependent transport protein |
| M. marinum M | MMAR_4137 | oppC | 9e-36 | 33.70% (276) | oligopeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_1432 | oppC | 2e-34 | 30.88% (285) | ABC transporter, permease protein OppC |
| M. smegmatis MC2 155 | MSMEG_4356 | - | 1e-44 | 37.82% (275) | inner membrane ABC transporter permease protein YddQ |
| M. thermoresistible (build 8) | TH_2705 | - | 6e-49 | 40.65% (278) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4000 | oppC | 6e-36 | 33.70% (276) | oligopeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 3e-49 | 40.71% (280) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1313c|M.bovis_AF2122/97 -----------------MTEFASRRTLVVRRFLRNRAAVASLAALLLLFV
Rv1282c|M.tuberculosis_H37Rv -----------------MTEFASRRTLVVRRFLRNRAAVASLAALLLLFV
MMAR_4137|M.marinum_M MTDQA-----------TQVEFTSRRTLVLRRFLRNRAAVASLAVLVVLFV
MUL_4000|M.ulcerans_Agy99 MTDQA-----------TQVEFTSRRTLVLRRFLRNRAAVASLAVLVVLFV
MAV_1432|M.avium_104 MTAQANVDGAHGFSGHEVAGFASRRTLVLRRFGRNRLAVTSLTLLVLLFV
MLBr_01123|M.leprae_Br4923 MTLTEG------LSISEKTNFASRKTLVLRRFAHNRTASVSLMLLVLLFV
Mflv_2759|M.gilvum_PYR-GCK MTTTDP---------------QSTRLSSWRLLALNPVTLISALVLIAVAV
Mvan_3775|M.vanbaalenii_PYR-1 MTTTDP---------------NDTRLSSWRLLAGNPVTLVSALVLVAVTV
MSMEG_4356|M.smegmatis_MC2_155 MTVT-----------------EQTRVASWRLLLSNPVTAVSAAVLVVVAV
TH_2705|M.thermoresistible__bu MTADDLT---------GGEPDAVSRVAAWRLLAANPVTLLSAALLALLVA
MAB_0721|M.abscessus_ATCC_1997 MTTSVSKPHG-----NRLRRLPGADLLRSVHGLQRWTLVVGLVLVAGFIA
. . : . .
Mb1313c|M.bovis_AF2122/97 SAYALPPLLPYSYDDLDFN----ALLQPPGTKHWLGTNALGQDLLAQTLR
Rv1282c|M.tuberculosis_H37Rv SAYALPPLLPYSYDDLDFN----ALLQPPGTKHWLGTNALGQDLLAQTLR
MMAR_4137|M.marinum_M GCYALPSVLPYSYEDLDFD----ALLQPPSTRHWLGTNALGQDLLAQTLR
MUL_4000|M.ulcerans_Agy99 GCYALPSVLPYSYEDLDFD----ALLQPPSTRHWLGTNALGQDLLAQTLR
MAV_1432|M.avium_104 GCYTLPAVLPYSYQDLDFD----ALLQPPNARHWLGTNALGQDLLAQILR
MLBr_01123|M.leprae_Br4923 ACYLLPSLLPYSYDDLDFN----ALLQPPNTRHWLGTNALGQDLLAQTLR
Mflv_2759|M.gilvum_PYR-GCK VAVTANWIIPYGVNDVDVP----NALAPPSGSHWFGTDELGRDVLSRVLV
Mvan_3775|M.vanbaalenii_PYR-1 VAVTANWLAPYGVNDVDVP----NALAPPSASHWFGTDELGRDVLSRVLV
MSMEG_4356|M.smegmatis_MC2_155 VALTAQWIAPFGVNDIDVP----SALQGPGGTHWFGTDELGRDVFSRILV
TH_2705|M.thermoresistible__bu VAVTAPWLAPYGVNDINVP----EALQPPSAAHWFGTDELGRDVLSRVLV
MAB_0721|M.abscessus_ATCC_1997 IAVLAPVLAPYGFSQTSADGLDFVRQQAPSWQHWFGTSVRGEDVLSRVLF
. : *:. .: . *. **:**. *.*:::: *
Mb1313c|M.bovis_AF2122/97 GMQKSMLIGVCVAVISTGIAATVGAISGYFGGWRDRTLMWVVDLLLVVPS
Rv1282c|M.tuberculosis_H37Rv GMQKSMLIGVCVAVISTGIAATVGAISGYFGGWRDRTLMWVVDLLLVVPS
MMAR_4137|M.marinum_M GMQKSMLIGVFVAVISTGIAATVGAVSGYFGGWRDRALMWLVDLLLVVPS
MUL_4000|M.ulcerans_Agy99 GMQKSMLIGVFVAVISTGIAATVGAVSGYFGGWRDRALMWLVDLLLVVPS
MAV_1432|M.avium_104 GMQKSMLIGVCVAVISTGIAATVGSIAGYFGGWRDRVLMWLVDLLLVVPS
MLBr_01123|M.leprae_Br4923 GMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLLVVPS
Mflv_2759|M.gilvum_PYR-GCK ATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRVVDVMFAFPV
Mvan_3775|M.vanbaalenii_PYR-1 AVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRVVDVMFAFPV
MSMEG_4356|M.smegmatis_MC2_155 AIQASLRVAVVSVALAAVVGVTIGVVAGYRGGWLDTIVMRVVDVMFAFPV
TH_2705|M.thermoresistible__bu AIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRLVDVLFAFPV
MAB_0721|M.abscessus_ATCC_1997 GARTALLVIVTSLVVSLPAGVVLGLTSGYLGGWLDRALVLVMDALYAMPS
. : :: : . .: . :* :** * * * :: ::* : ..*
Mb1313c|M.bovis_AF2122/97 FILIAIVTPRTKNSAN-----IMFLVLLLAGFGWMISSRMVRGMTMSLRE
Rv1282c|M.tuberculosis_H37Rv FILIAIVTPRTKNSAN-----IMFLVLLLAGFGWMISSRMVRGMTMSLRE
MMAR_4137|M.marinum_M FILIAIVTPRTKNSTN-----MMFLVLLLAGFGWMISSRMVRGMTMSLRE
MUL_4000|M.ulcerans_Agy99 FILIAIVTPRTKNSTN-----MMFLVLLLAGFGWMISSRMVRGMTMSLRE
MAV_1432|M.avium_104 FILIAIVTPRTKNSAN-----ILMLVLLLAGFGWMVSSRMVRGMTMSLRE
MLBr_01123|M.leprae_Br4923 FVLIAIMSPQTKSSDN-----IMLLVLLLAGFGWMVSSRMVRGMTMSLRK
Mflv_2759|M.gilvum_PYR-GCK LLLALAVVAILGPGVT-----TTILAIGIVYT--PIFARVARASTLGVRT
Mvan_3775|M.vanbaalenii_PYR-1 LLLALAVVAILGPGVT-----TTILAIAIVYT--PIFARVARASTLGVRT
MSMEG_4356|M.smegmatis_MC2_155 LLLALAIVAILGPGVT-----TTMLAIGIVYI--PIFARVARASTLGVRV
TH_2705|M.thermoresistible__bu LLLALAIVAILGPGMT-----TTMLAIGIVFT--PIFARVARAGTLGVRV
MAB_0721|M.abscessus_ATCC_1997 LLLAIVVSIAVTGGQSSTGGGILAAAIAIMAVFVPQYFRVVRNATVGVKQ
::* : . . .: : *:.* *:.::
Mb1313c|M.bovis_AF2122/97 REFIRAARYMGVSSRRIIVGHVVPNVASILIIDAALNVAAAILAETGLSF
Rv1282c|M.tuberculosis_H37Rv REFIRAARYMGVSSRRIIVGHVVPNVASILIIDAALNVAAAILAETGLSF
MMAR_4137|M.marinum_M REFIRAARYMGVSSRRIITGHVIPNVASILIIDAALNVAAAILAETGLSF
MUL_4000|M.ulcerans_Agy99 REFIRAARYMGVSSRRIITGHVIPNVASILIIDAALNVAAAILAETGLSF
MAV_1432|M.avium_104 REFIRAARYMGVSSRRIIVGHVVPNVASILIIDAALNVASAILAETGLSF
MLBr_01123|M.leprae_Br4923 SEFIRAGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSF
Mflv_2759|M.gilvum_PYR-GCK EPYVSVSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSF
Mvan_3775|M.vanbaalenii_PYR-1 EPYVSASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSF
MSMEG_4356|M.smegmatis_MC2_155 EPYVAVSRTMGTPSGFILRRHILPNIAGPLIVQLSLSLAFAILAEASLSF
TH_2705|M.thermoresistible__bu EPFVQASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFAILAEAALSF
MAB_0721|M.abscessus_ATCC_1997 EPFVDAARVTGAGTPRILFRHILSNVTSSLPVIITLNGAEAILTLAGLGF
:: ..: *. :: *::.*::. * : :*. . ***: :.*.*
Mb1313c|M.bovis_AF2122/97 LGFGIQPPDVSLG--TLIADGTASATAFPWVFLFPASILVLILVCANLTG
Rv1282c|M.tuberculosis_H37Rv LGFGIQPPDVSLG--TLIADGTASATAFPWVFLFPASILVLILVCANLTG
MMAR_4137|M.marinum_M LGFGIQPPDVSLG--TLIADGTQSATTFPWVFLFPAGVLVLILVCANLTG
MUL_4000|M.ulcerans_Agy99 LGFGIQPPDVSLG--TLIADGTQSATTFPWVFLFPAGVLVLILVCANLTG
MAV_1432|M.avium_104 LGFGVQPPDVSLG--TLIANGTQSATTFPWVFLFPAGVLVLILVCANLTG
MLBr_01123|M.leprae_Br4923 LGFGIKPPDVSLG--TLIANGTQSATTFPWVFLFPAGVLVVILVCANLTG
Mflv_2759|M.gilvum_PYR-GCK LGLGIQPPQPSLG--RMIFDSQGFVTMAWWMAVFPGAAIFLIVLAFNLFG
Mvan_3775|M.vanbaalenii_PYR-1 LGLGIQPPQPSLG--RMIFDSQGFVTMAWWMAVFPGAAIFVIVLAFNLLG
MSMEG_4356|M.smegmatis_MC2_155 LGLGIQPPQPSLG--RMIFDAQGFVTLAWWMAVFPGAAIFVTVLAFNLFG
TH_2705|M.thermoresistible__bu LGLGIQPPEPSLG--RMIFDSQGFVTLAWWMAVFPGAAIVAAVLACNLFG
MAB_0721|M.abscessus_ATCC_1997 LGFGIEPTQAAEWGYDLNKALSDVANGIWWTGVFPGIAIVLVVLGMTLVG
**:*::*.: : : .. * :**. :. :: .* *
Mb1313c|M.bovis_AF2122/97 DGLRDALDPASRSLRRGVR----------------
Rv1282c|M.tuberculosis_H37Rv DGLRDALDPASRSLRRGVR----------------
MMAR_4137|M.marinum_M DGLRDALDPASKTLRSGDR----------------
MUL_4000|M.ulcerans_Agy99 DGLRDALDPASKTLRSGDR----------------
MAV_1432|M.avium_104 DGLRDALDPGSGPARGGRR----------------
MLBr_01123|M.leprae_Br4923 DGLRDAFDPVSKHLRR-------------------
Mflv_2759|M.gilvum_PYR-GCK DGLRDVLDPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 DGLRDVLDPKQRTTVEARRRER-------------
MSMEG_4356|M.smegmatis_MC2_155 DGLRDVLDPKQRTLMEAKRTRQ-------------
TH_2705|M.thermoresistible__bu DGLRDVLDPKQRTVIETQRRARQRPGRGRRAGSAW
MAB_0721|M.abscessus_ATCC_1997 ESLNEVINPLLRTRRGEA-----------------
:.*.:.::*