For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVMVKAVLFDFSGTLFRLEEDESWFEGMTVDEREVDGHVQAELMRRLTAPTGRSVQMSDEQYHAWVNRDL APHLHREAYLHVLRESGLADPHAESLYGRVIDPDSWTAYPDTATVLKSLKQQDIKTAVVSNIAFDVRPAF TALGIDADVDEFVLSYEVGAVKPDAAIFTTTLQRLGVAPQDALMVGDSEEADGGATAVGCAFALVDPLPT TERPDGLIAAVSSVGIAT*
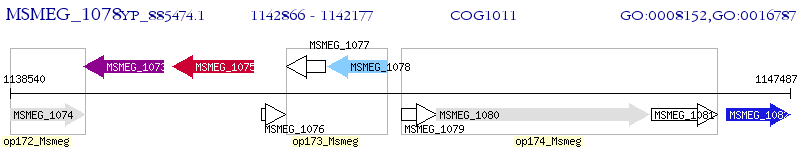
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1078 | - | - | 100% (229) | hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3410 | - | 6e-07 | 27.73% (119) | hypothetical protein Mb3410 |
| M. gilvum PYR-GCK | Mflv_0023 | - | 8e-90 | 72.93% (229) | HAD family hydrolase |
| M. tuberculosis H37Rv | Rv3376 | - | 6e-07 | 27.73% (119) | hypothetical protein Rv3376 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_4652 | - | 2e-12 | 80.00% (40) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0949 | - | 5e-91 | 73.04% (230) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0023|M.gilvum_PYR-GCK -MTDLDAVLFDFSGTLF-----RLEEDESWFDGMTLDS----AGNRDVDE
Mvan_0949|M.vanbaalenii_PYR-1 -MSDLQAVLFDYSGTLF-----RLEEDESWFAGMELDTPLSPDANREIDG
MSMEG_1078|M.smegmatis_MC2_155 MVVMVKAVLFDFSGTLF-----RLEEDESWFEGMTVDE-------REVDG
TH_4652|M.thermoresistible__bu -------VLFDFSGTLF-----RLEEDASWFGGMTIAE------QREVDR
Mb3410|M.bovis_AF2122/97 --MSISAVVFDRDGVLTSFDWTRAEEDVRRITGLPLEE-----IERRWGG
Rv3376|M.tuberculosis_H37Rv --MSISAVVFDRDGVLTSFDWTRAEEDVRRITGLPLEE-----IERRWGG
*:** .*.* * *** : *: : * .
Mflv_0023|M.gilvum_PYR-GCK HVQAELMRRLTAPTGRSVSMTPEALEAWMKRDLAPHLHREAYLHVLRESG
Mvan_0949|M.vanbaalenii_PYR-1 HVQAELMRRLTAPTGHSVSMTPDALDAWLNRDLAPHLHREAYLHVLRESG
MSMEG_1078|M.smegmatis_MC2_155 HVQAELMRRLTAPTGRSVQMSDEQYHAWVNRDLAPHLHREAYLHVLRESG
TH_4652|M.thermoresistible__bu HVQVELMR------------------------------------------
Mb3410|M.bovis_AF2122/97 WLNGLTIDDAFVETQP----------------------ISEFLSSLAREL
Rv3376|M.tuberculosis_H37Rv WLNGLTIDDAFVETQP----------------------ISEFLSSLAREL
:: :
Mflv_0023|M.gilvum_PYR-GCK LACEHAESLYSRVIDPACWTPYPDTATVLTGLRRRGIRTAVVSN--IAFD
Mvan_0949|M.vanbaalenii_PYR-1 LAHHHAESLYSRVIDPACWTPYPDTATVLDGLHRRGISTAVVSN--IAFD
MSMEG_1078|M.smegmatis_MC2_155 LADPHAESLYGRVIDPDSWTAYPDTATVLKSLKQQDIKTAVVSN--IAFD
TH_4652|M.thermoresistible__bu --------------------------------------------------
Mb3410|M.bovis_AF2122/97 ELGSKARDELVRLDYMAFAQGYPDARPALEEARRRGLKVGVLTNNSLLVS
Rv3376|M.tuberculosis_H37Rv ELGSKARDELVRLDYMAFAQGYPDARPALEEARRRGLKVGVLTNNSLLVS
Mflv_0023|M.gilvum_PYR-GCK LRPAFDGVG---TVDEFVLSFEVGAVKPDPSIFQTALDRLGVPAERALMV
Mvan_0949|M.vanbaalenii_PYR-1 VRPAFEVLGAAGHVDEFVLSFEVGAVKPDPAIFQTALDRLGVAADRALMV
MSMEG_1078|M.smegmatis_MC2_155 VRPAFTALGIDADVDEFVLSYEVGAVKPDAAIFTTTLQRLGVAPQDALMV
TH_4652|M.thermoresistible__bu --------------------------------------------------
Mb3410|M.bovis_AF2122/97 ARSLLQCAALHDLVDVVLSSQMIGAAKPDPRAYQAIAEALGVSTTSCLFF
Rv3376|M.tuberculosis_H37Rv ARSLLQCAALHDLVDVVLSSQMIGAAKPDPRAYQAIAEALGVSTTSCLFF
Mflv_0023|M.gilvum_PYR-GCK GDSDEADGGARALGCAFALVDPLPTRERSDGLLGALRAHGIRL-----
Mvan_0949|M.vanbaalenii_PYR-1 GDSDEADGGARTLGCAFALVDPLPTAERKDGLVAALWANGIQL-----
MSMEG_1078|M.smegmatis_MC2_155 GDSEEADGGATAVGCAFALVDPLPTTERPDGLIAAVSSVGIAT-----
TH_4652|M.thermoresistible__bu ----------------LVLVDPLPSGQRPDGLRTALRDHGLRV-----
Mb3410|M.bovis_AF2122/97 DDIADWVEGARCAGMRAYLVD--RSGQTRDGVVRDLSSLGAILDGAGP
Rv3376|M.tuberculosis_H37Rv DDIADWVEGARCAGMRAYLVD--RSGQTRDGVVRDLSSLGAILDGAGP
*** : : **: : *