For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRISTVVGVLAAGVLIGGVAAPSAAAQSTCAELGGTVDADQICQVHTANATYRLDYTFSTTYPDQEAVVG YLSQTRDGFVNVSDMPGSRDQPYVLDARGSSYSSAVAPGLPARTQSLVLEVFQDVGGAQPETWYKSFNYD LVKRAPITFDTLFKPGSRPLDVIFPIVARELETQAGIVQSISQSAGMDPTHYQNFAITDDSVIFFFDRGA LVADSAGALQASVPRWAVASLLA
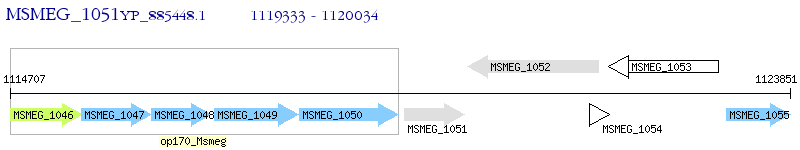
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1051 | - | - | 100% (233) | immunogenic protein MPB64/MPT64 |
| M. smegmatis MC2 155 | MSMEG_2331 | - | e-131 | 100.00% (233) | immunogenic protein MPB64/MPT64 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3062c | TB22.2 | 3e-64 | 53.68% (231) | hypothetical protein Mb3062c |
| M. gilvum PYR-GCK | Mflv_4267 | - | 4e-67 | 53.91% (230) | immunogenic protein MPB64/MPT64 precursor |
| M. tuberculosis H37Rv | Rv3036c | TB22.2 | 2e-64 | 53.68% (231) | hypothetical protein Rv3036c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1835c | - | 6e-30 | 31.14% (273) | immunogenic protein MPT64 precursor |
| M. marinum M | MMAR_1665 | - | 2e-62 | 52.00% (225) | hypothetical protein MMAR_1665 |
| M. avium 104 | MAV_3901 | - | 7e-56 | 45.89% (231) | immunogenic protein MPB64/MPT64 |
| M. thermoresistible (build 8) | TH_2187 | - | 4e-67 | 55.17% (232) | PROBABLE CONSERVED SECRETED PROTEIN TB22.2 |
| M. ulcerans Agy99 | MUL_1902 | - | 4e-61 | 50.44% (228) | hypothetical protein MUL_1902 |
| M. vanbaalenii PYR-1 | Mvan_2092 | - | 8e-75 | 58.33% (228) | immunogenic protein MPB64/MPT64 precursor |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4267|M.gilvum_PYR-GCK MRFLAAALTAAAVSASLAAGPAALAQPAPPA-TPSACVALSGTVDTDDMC
Mvan_2092|M.vanbaalenii_PYR-1 MRIFAAALTAAGATAGLLAVPAAGLLAAPAAGAQSACAELGGTVDAEQTC
TH_2187|M.thermoresistible__bu MRILKT-------TAALLAGGLISCSIPGTAGAQTACDALGGTVADDRRC
MSMEG_1051|M.smegmatis_MC2_155 MRISTV-------VGVLAAGVLIGGVAAPSAAAQSTCAELGGTVDADQIC
Mb3062c|M.bovis_AF2122/97 MRYLIA-------TAVLVAVVLVG---WPAAGAPPSCAGLGGTVQAGQIC
Rv3036c|M.tuberculosis_H37Rv MRYLIA-------TAVLVAVVLVG---WPAAGAPPSCAGLGGTVQAGQIC
MMAR_1665|M.marinum_M ---MIA-------AAMLVSAVLLA---WPAAAEPPSCASLGGNVEGAQMC
MUL_1902|M.ulcerans_Agy99 MRLMIA-------AAMLVSAVLLA---WPAAAEPPSCASLGGNVEGAQMC
MAV_3901|M.avium_104 MRYLLA-------AALLAPAVLLG---WPAAAEPPSCAALGGSLEDGQMC
MAB_1835c|M.abscessus_ATCC_199 MPYLSLR--SISASVLVTAASIFGFSGVAGAAPHDYCGDLKGTN-TGKTC
: . * * * *. *
Mflv_4267|M.gilvum_PYR-GCK LVQVENPTYRLDYSFPADYPDQQALTAYLTQTRDGFVNVAGMPGSWNLPY
Mvan_2092|M.vanbaalenii_PYR-1 HVHAENSTYRLDYTFPVDYPDQQALAAYLTQTRDGFVNVSEMPGSWNLPY
TH_2187|M.thermoresistible__bu EVHAETPTYLLDMVFPVDYPDQQALTDYLIQTRDGFVNVSEMPGSRNLPY
MSMEG_1051|M.smegmatis_MC2_155 QVHTANATYRLDYTFSTTYPDQEAVVGYLSQTRDGFVNVSDMPGSRDQPY
Mb3062c|M.bovis_AF2122/97 HVHASGPKYMLDMTFPVDYPDQRALTDYITQNRDGFVNVAQGSPLRDQPY
Rv3036c|M.tuberculosis_H37Rv HVHASGPKYMLDMTFPVDYPDQQALTDYITQNRDGFVNVAQGSPLRDQPY
MMAR_1665|M.marinum_M HIHATGSTYTLDITFPQDYPDQQALTDYITQNRDGFVSVATGSGRRDQPY
MUL_1902|M.ulcerans_Agy99 HIHATGSTYTLDITFPQDYPDQQALTDYITLNRDGFVSVATGSGRRDQPY
MAV_3901|M.avium_104 RLRAAGPNYTINMVFPADYPDEQALTDYITQNRDGFVNVAQSSGGRDQPY
MAB_1835c|M.abscessus_ATCC_199 EIRLSDTGYSVDVTIPLNYPDQKSIAEYVAKTRDAFVNAAKSGAARSTTA
:: . * :: :. ***:.::. *: .**.**..: . .
Mflv_4267|M.gilvum_PYR-GCK VLDGRGTGYRTGPDD---GGTRSVVFEMYENVGGAHPQTWFKSFNWDVAK
Mvan_2092|M.vanbaalenii_PYR-1 VLDGRGTGYRTGPDG---AGTRSVVFEVYENVGGAHPQTWFKAFNWDVAK
TH_2187|M.thermoresistible__bu ALDARATEYRSGDAA---TGTRSVVFEVYQNVGGAHPTTWYQAFNYDLRT
MSMEG_1051|M.smegmatis_MC2_155 VLDARGSSYSSAVAPGLPARTQSLVLEVFQDVGGAQPETWYKSFNYDLVK
Mb3062c|M.bovis_AF2122/97 QMDATSEQHSSGQPP---QATRSVVLKFFQDLGGAHPSTWYKAFNYNLAT
Rv3036c|M.tuberculosis_H37Rv QMDATSEQHSSGQPP---QATRSVVLKFFQDLGGAHPSTWYKAFNYNLAT
MMAR_1665|M.marinum_M QMDATTEQHAAGQPP---HNTRSVVLKFFQDLGGAHPFTWYKAFNYNLGT
MUL_1902|M.ulcerans_Agy99 QMDATTEQHAAGQPP---HNTRSVVLKFFQDLGGAHPFTWHKAFNYNLGT
MAV_3901|M.avium_104 QLEATTEQHSAGQPP---HNTRSVVLKFFQDVGGTRSSIWYKAFNYNLGT
MAB_1835c|M.abscessus_ATCC_199 QLSMKPTEYNSDLPP---RGTQTVVFKVYQTGGNGQPQTAYKAFNWDQSY
:. : : *:::*::.:: *. :. .::**::
Mflv_4267|M.gilvum_PYR-GCK KAPITFDTLFKPGS--------EPLAVIFPIVRSDISRQLG---------
Mvan_2092|M.vanbaalenii_PYR-1 KAPITFDTLFRPDT--------KPLDVIFPIVQKDISRQLG---------
TH_2187|M.thermoresistible__bu RAPITFDTLFDPES--------APLEVIYPVVQRELQELTG---------
MSMEG_1051|M.smegmatis_MC2_155 RAPITFDTLFKPGS--------RPLDVIFPIVARELETQAG---------
Mb3062c|M.bovis_AF2122/97 SQPITFDTLFVPGT--------TPLDSIYPIVQRELARQTG---------
Rv3036c|M.tuberculosis_H37Rv SQPITFDTLFVPGT--------TPLDSIYPIVQRELARQTG---------
MMAR_1665|M.marinum_M KQPITFDTLFAPGT--------TPLDAIYPIVQRELAHQTG---------
MUL_1902|M.ulcerans_Agy99 KQPITFDTLFAPGT--------TPLDAIYPIVQRELAHQTG---------
MAV_3901|M.avium_104 KQPLTFDNLFAPNT--------APLDAIFPIVQRDLERQTP---------
MAB_1835c|M.abscessus_ATCC_199 RKPVLYTVPKDDKDDAPLWRVDDPLKTVAPIVQAQLQQQLAPPPVATPTP
*: : ** : *:* ::
Mflv_4267|M.gilvum_PYR-GCK ------------------------IEAPITTADGLDPAKYTEFAITDDAV
Mvan_2092|M.vanbaalenii_PYR-1 ------------------------VDAPISAADGLDPAKYQEFALTDDSL
TH_2187|M.thermoresistible__bu ------------------------IDQPILPEDGLDPANYQNFALTDDEV
MSMEG_1051|M.smegmatis_MC2_155 ------------------------IVQSISQSAGMDPTHYQNFAITDDSV
Mb3062c|M.bovis_AF2122/97 ------------------------FGAAILPSTGLDPAHYQNFAITDDSL
Rv3036c|M.tuberculosis_H37Rv ------------------------FGAAILPSTGLDPAHYQNFAITDDSL
MMAR_1665|M.marinum_M ------------------------LGVAILPSNGLDPTHYQNFAITDTDV
MUL_1902|M.ulcerans_Agy99 ------------------------LGVAILPSNGLDPTHYQNFAITDTDV
MAV_3901|M.avium_104 ------------------------LGAAILPSTGHDPSHYQNFAITDDQL
MAB_1835c|M.abscessus_ATCC_199 TTTSGQPTTTTSTTTTSTTPPPPPPPLPFSPATLYDPANYQNFAVLNDGI
.: **::* :**: : :
Mflv_4267|M.gilvum_PYR-GCK IFYFGQGEIMAGAGGALQATIPRSEIAS-------------MLALEPVPV
Mvan_2092|M.vanbaalenii_PYR-1 IFYFGQGEIMPGAGGALQATVPRSAVAA-------------MLAEPAVTQ
TH_2187|M.thermoresistible__bu IFFFSQGELLPHAAGVSQVSVPRTVLVN-------------LLTV-----
MSMEG_1051|M.smegmatis_MC2_155 IFFFDRGALVADSAGALQASVPRWAVAS-------------LLA------
Mb3062c|M.bovis_AF2122/97 IFYFAQGELLPSFVGACQAQVPRSAIPP--------------LAI-----
Rv3036c|M.tuberculosis_H37Rv IFYFAQGELLPSFVGACQAQVPRSAIPP--------------LAI-----
MMAR_1665|M.marinum_M IFYFAQGELLPSFAGATQAQVPRNAIPP--------------LAI-----
MUL_1902|M.ulcerans_Agy99 IFYFAQGELLPSFAGATQAQVPRNAIPPPWRSEVHTGPASDSIAAAPMPS
MAV_3901|M.avium_104 IFYFAPGEMLPAFGGPVQAQVPRNAIPP--------------LAI-----
MAB_1835c|M.abscessus_ATCC_199 RFFFDQGAILPDSYGALQVLVPRSAIDP-------------MIA------
*:* * ::. * *. :** : ::
Mflv_4267|M.gilvum_PYR-GCK TP------------------------------------------------
Mvan_2092|M.vanbaalenii_PYR-1 G-------------------------------------------------
TH_2187|M.thermoresistible__bu --------------------------------------------------
MSMEG_1051|M.smegmatis_MC2_155 --------------------------------------------------
Mb3062c|M.bovis_AF2122/97 --------------------------------------------------
Rv3036c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1665|M.marinum_M --------------------------------------------------
MUL_1902|M.ulcerans_Agy99 VSWATHPMIGTTKATLCRASRYTLMMRSARNITMPPQKRSPIALSTQEGS
MAV_3901|M.avium_104 --------------------------------------------------
MAB_1835c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4267|M.gilvum_PYR-GCK ----
Mvan_2092|M.vanbaalenii_PYR-1 ----
TH_2187|M.thermoresistible__bu ----
MSMEG_1051|M.smegmatis_MC2_155 ----
Mb3062c|M.bovis_AF2122/97 ----
Rv3036c|M.tuberculosis_H37Rv ----
MMAR_1665|M.marinum_M ----
MUL_1902|M.ulcerans_Agy99 PQVW
MAV_3901|M.avium_104 ----
MAB_1835c|M.abscessus_ATCC_199 ----