For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RETLTPNFADVQAHYDLSDDFFRLFLDPTQTYSCAYFAQDGMTLEEAQIAKIDLALGKLGLEPGMTLLDV GCGWGATMRRAVERHDVNVVGLTLSRNQAAHVERMLDDVGSRRSRRVLLQGWEQFDEPVDRIVSIGAFEH FRHERYDEFFAKAHALLPDDGVMLLHTICGFHPNHFREAGIPMTMEMAKFVKFILTEIFPGGQLPSSEMV HDRAGRAGFVVSREQSLQPHYAKTLDIWASRLQAHQDEAIAIQSLEVYQRYMKYLTGCAQLFRDGQVDVV QFTLQKGR*
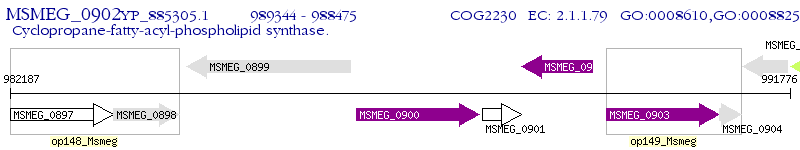
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0902 | - | - | 100% (289) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_1351 | - | e-121 | 71.73% (283) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_3538 | - | e-116 | 69.96% (283) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3424c | cmaA1 | 1e-122 | 72.98% (285) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
| M. gilvum PYR-GCK | Mflv_3304 | - | 1e-112 | 68.55% (283) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. tuberculosis H37Rv | Rv3392c | cmaA1 | 1e-122 | 72.63% (285) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
| M. leprae Br4923 | MLBr_02459 | umaA2 | 1e-111 | 68.20% (283) | Mycolic acid synthase |
| M. abscessus ATCC 19977 | MAB_3881 | - | 1e-112 | 67.14% (283) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. marinum M | MMAR_2920 | mmaA2_1 | 1e-123 | 73.24% (284) | methoxy mycolic acid synthase 2 MmaA2 _1 |
| M. avium 104 | MAV_0130 | - | 1e-116 | 70.77% (284) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. thermoresistible (build 8) | TH_4640 | - | 1e-121 | 73.14% (283) | PUTATIVE methoxy mycolic acid synthase 5 MmaA5_1 |
| M. ulcerans Agy99 | MUL_3772 | mmaA5 | 1e-120 | 71.23% (285) | methoxy mycolic acid synthase 5 MmaA5 |
| M. vanbaalenii PYR-1 | Mvan_3025 | - | 1e-114 | 68.55% (283) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3304|M.gilvum_PYR-GCK MSDTPNGSADLQPHFDDVQSHYDLSDDFYRLFLDPTQTYSCAYFERDDMT
Mvan_3025|M.vanbaalenii_PYR-1 MPETTSGSADLQPHFEDVQSHYDLSDDFYRLFLDPTQTYSCAYFERDDMT
MAV_0130|M.avium_104 MSENTGDAVNLQPHFDDVQAHYDLSDEFFALFLDPTRTYSCAYFERDDMT
MLBr_02459|M.leprae_Br4923 ------MSVQLTPHFRNVQAHYDLSDDFFRLFLDPTLTYSCAYFERDDMT
MMAR_2920|M.marinum_M ------MAQRLVPHFDDVQAHYDLSDDFFRLFLDPTQTYSCAYFERDDMT
MUL_3772|M.ulcerans_Agy99 ------MAKHLAPRFDDVQAHYDLSDDFFRLFLDPTQTYSCAYFERDDMT
TH_4640|M.thermoresistible__bu ---MAEPAKKLKPHFADVQAHYDLSDEFFGLFLDPTRTYSCAYFERDDMS
Mb3424c|M.bovis_AF2122/97 ------MPDELKPHFANVQAHYDLSDDFFRLFLDPTQTYSCAYFERDDMT
Rv3392c|M.tuberculosis_H37Rv ------MPDELKPHFANVQAHYDLSDDFFRLFLDPTQTYSCAYFERDDMT
MSMEG_0902|M.smegmatis_MC2_155 ------MRETLTPNFADVQAHYDLSDDFFRLFLDPTQTYSCAYFAQDGMT
MAB_3881|M.abscessus_ATCC_1997 -------MSKLTPKFSDVQAHYDLSDDFFALFLDPSRTYSCAYFEPETLT
* *.* :**:******:*: *****: ******* : ::
Mflv_3304|M.gilvum_PYR-GCK LEEAQRAKIDLALGKLGLEPGMTLLDIGCGWGGTLLRALEKYDVNVIGLT
Mvan_3025|M.vanbaalenii_PYR-1 LEEAQIAKIDLSLGKLGLQPGMTLLDIGCGWGATLLRALDKYDVNVIGLT
MAV_0130|M.avium_104 LEEAQIAKVDLSLGKLGLQPGMTLLDIGCGWGTTIVRALERYDVNVVGLT
MLBr_02459|M.leprae_Br4923 LEEAQLAKVDLALGKLKLEPGMTLLDIGCGWGATMKRAIEIYDVNVIGLT
MMAR_2920|M.marinum_M LEEAQIAKIDLALGKLGLEPGMTLLDIGCGWGATMRRAIEKHDVNVVGLT
MUL_3772|M.ulcerans_Agy99 LEEAQIAKIDLALGKLGLEPGMTLLDIGCGWGATMRRAIEKYDVNVVGLT
TH_4640|M.thermoresistible__bu LEEAQLAKIDLALGKLGLQPGMTLLDIGCGWGSTMLRALERHDVDVVGLT
Mb3424c|M.bovis_AF2122/97 LQEAQIAKIDLALGKLGLQPGMTLLDVGCGWGATMMRAVEKYDVNVVGLT
Rv3392c|M.tuberculosis_H37Rv LQEAQIAKIDLALGKLGLQPGMTLLDVGCGWGATMMRAVEKYDVNVVGLT
MSMEG_0902|M.smegmatis_MC2_155 LEEAQIAKIDLALGKLGLEPGMTLLDVGCGWGATMRRAVERHDVNVVGLT
MAB_3881|M.abscessus_ATCC_1997 LEQAQQAKIDLALGKLNLEPGMTLLDVGCGWGSTMKRALEHYDVNVIGLT
*::** **:**:**** *:*******:***** *: **:: :**:*:***
Mflv_3304|M.gilvum_PYR-GCK LSRNQQAHVQKVLDEHDSPRSKTVLLQGWEQFDQPVDRIVSIGAFEHFGS
Mvan_3025|M.vanbaalenii_PYR-1 LSRNQQAHVQKRLDAHGSGRSKRVLLAGWEQFDEPVDRIVSIGAFEHFGR
MAV_0130|M.avium_104 LSRNQQAHVQRPLDNHPSPRSKRVLLQGWEQFDEKVDRIVSIGAFEHFGR
MLBr_02459|M.leprae_Br4923 LSENQAAHVQKTFDELDTPRTRRVLLEGWEKFHEPVDRIVSIGAFEHFGR
MMAR_2920|M.marinum_M LSNNQAAHVQKSFDQLDTARTRRVLLKGWEQFDEPVDRIVSIGAFEHFGH
MUL_3772|M.ulcerans_Agy99 LSKNQAAHVQKSFDQLDTARTRRVLLEGWEQFDEPVDRIVSIGAFEHFGH
TH_4640|M.thermoresistible__bu LSANQHAHVEQLLADSPSRRSRRVLLRGWEEFDEPVDRIVSIGAFEHFGH
Mb3424c|M.bovis_AF2122/97 LSKNQANHVQQLVANSESLRSKRVLLAGWEQFDEPVDRIVSIGAFEHFGH
Rv3392c|M.tuberculosis_H37Rv LSKNQANHVQQLVANSENLRSKRVLLAGWEQFDEPVDRIVSIGAFEHFGH
MSMEG_0902|M.smegmatis_MC2_155 LSRNQAAHVERMLDDVGSRRSRRVLLQGWEQFDEPVDRIVSIGAFEHFRH
MAB_3881|M.abscessus_ATCC_1997 LSKNQFAYVNTLLDSAGSSRNYEVRLAGWEEFEGTVDRIVSIGAFEHFGH
** ** :*: . . *. * * ***:*. *************
Mflv_3304|M.gilvum_PYR-GCK DRYDEFFRRAYSVLPDDGVMLLHTIVKPSDEEFAERKLPLTMTKLKFFKF
Mvan_3025|M.vanbaalenii_PYR-1 DRYDEFFKKAYQVLPDGGVMMLHTIIKPTDEEFADRGLKLTMSIVRFSKF
MAV_0130|M.avium_104 DRYDDFFKMAYAALPDDGVMMLHTIIKPSDEEFAERGLPITMTKLRFMKF
MLBr_02459|M.leprae_Br4923 QRYARFFKMAHEVLPADSVMLLHTIVRPTFKDARARGMTLTHEIVHFTQF
MMAR_2920|M.marinum_M DRYDDFFAMAHRVLPRGGVMLLHTITGLTREQISDRGIPISIDMIRFVKF
MUL_3772|M.ulcerans_Agy99 DRYDDFFTLAHNILPSDGVMLLHTITGLTMPQMVENGLPLTLWLARFLKF
TH_4640|M.thermoresistible__bu DRYDDFFATAHRLLPDDGVMLLHTITALTVPQMVDRGIPLTFEVARFVKF
Mb3424c|M.bovis_AF2122/97 ERYDAFFSLAHRLLPADGVMLLHTITGLHPKEIHERGLPMSFTFARFLKF
Rv3392c|M.tuberculosis_H37Rv ERYDAFFSLAHRLLPADGVMLLHTITGLHPKEIHERGLPMSFTFARFLKF
MSMEG_0902|M.smegmatis_MC2_155 ERYDEFFAKAHALLPDDGVMLLHTICGFHPNHFREAGIPMTMEMAKFVKF
MAB_3881|M.abscessus_ATCC_1997 ERYDSFFAMAHRVLPSDGKMLLHTIVSHHPDEWKRMGIPLLMSRVRFIYF
:** ** *: ** .. *:**** . : : :* *
Mflv_3304|M.gilvum_PYR-GCK IMDEIFPGGMLPKVDDVEKHATKASFKVNRIQPLRLHYARTLQIWAASLE
Mvan_3025|M.vanbaalenii_PYR-1 IMDEIFPGGDLPKPTTVAEHATKAGFRLTREQRLREHYAKTLDIWADALR
MAV_0130|M.avium_104 IMDEIFPGGDLPQAKGVIEHAQRAGFGVQRVQALRPHYARTLDTWAAALR
MLBr_02459|M.leprae_Br4923 ILAEIFPGGWLPTVPTVEEHATAAGFKFTRIQSLQLHYAKTLDLWAARLE
MMAR_2920|M.marinum_M IVTEIFPGGRLPSIDMVQQHAGRAGFTLTRRQSLQPHYARTLALWAQALY
MUL_3772|M.ulcerans_Agy99 IQTEIFPGGHPPTIEMVEEQSVKPGFTLTRRHSLQLHYAKTLDLWAEALE
TH_4640|M.thermoresistible__bu ILTEIFPGGRLPSIEKVEDHSGANGFTLTRRQSLQSHYARTLDLWAEALQ
Mb3424c|M.bovis_AF2122/97 IVTEIFPGGRLPSIPMVQECASANGFTVTRVQSLQPHYAKTLDLWSAALQ
Rv3392c|M.tuberculosis_H37Rv IVTEIFPGGRLPSIPMVQECASANGFTVTRVQSLQPHYAKTLDLWSAALQ
MSMEG_0902|M.smegmatis_MC2_155 ILTEIFPGGQLPSSEMVHDRAGRAGFVVSREQSLQPHYAKTLDIWASRLQ
MAB_3881|M.abscessus_ATCC_1997 IGTEIFPGGRLPSVPMVDKHAALAGFHITRHHSLQPHYARTLDCWATNLA
* ****** * * . : .* . * : *: ***:** *: *
Mflv_3304|M.gilvum_PYR-GCK SRRDDAIAIQSEEVYDRYMKYLTGCAELFTEGYTDVCQFTLAK-------
Mvan_3025|M.vanbaalenii_PYR-1 KREDDAVAIQSREDYDRYMKYLTGCADLFRDGYTDICQFTLEK-------
MAV_0130|M.avium_104 ARRDEAIAIQSEEVYDRYMKYLTGCADLFREGYTDVAQFTLTKG------
MLBr_02459|M.leprae_Br4923 ANKNKAIAIQSEEVYDRYMKYLTGCAKLFRDGYTDVDQFTLEKQPATARR
MMAR_2920|M.marinum_M AYRDEAIEIQSEEVYERYLKYLTGCADAFRRGHIDVNQFTLQN-------
MUL_3772|M.ulcerans_Agy99 VRKDEAIKIQSDEVYERYMRYLTGCSKLFQEGYVDVNQFTLQKAR-----
TH_4640|M.thermoresistible__bu QHREEAITVQSDEVYDRYIKYLTGCAKGFRVGYIDVNQFTLQK-------
Mb3424c|M.bovis_AF2122/97 ANKGQAIALQSEEVYERYMKYLTGCAEMFRIGYIDVNQFTCQK-------
Rv3392c|M.tuberculosis_H37Rv ANKGQAIALQSEEVYERYMKYLTGCAEMFRIGYIDVNQFTCQK-------
MSMEG_0902|M.smegmatis_MC2_155 AHQDEAIAIQSLEVYQRYMKYLTGCAQLFRDGQVDVVQFTLQKGR-----
MAB_3881|M.abscessus_ATCC_1997 AHEDEAISVQSQEVYDRYIKYLTGCAEGFREGWIDVVQFTCEK-------
. .*: :** * *:**::*****:. * * *: *** :
Mflv_3304|M.gilvum_PYR-GCK -
Mvan_3025|M.vanbaalenii_PYR-1 -
MAV_0130|M.avium_104 -
MLBr_02459|M.leprae_Br4923 H
MMAR_2920|M.marinum_M -
MUL_3772|M.ulcerans_Agy99 -
TH_4640|M.thermoresistible__bu -
Mb3424c|M.bovis_AF2122/97 -
Rv3392c|M.tuberculosis_H37Rv -
MSMEG_0902|M.smegmatis_MC2_155 -
MAB_3881|M.abscessus_ATCC_1997 -