For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ANTVLPTVERSTRVGDPERERTADDLGRALAQGYLGMPEYEERLQRVFGAQTTAQLRDLVADLPVAALRR NDPRRREAQMRAARMSVRLHLAAYVAGSVLMLGIWLAVGLGGGGWYFWPVWPIMGWGIGVASHAIPIWAH GSMRPARIA*
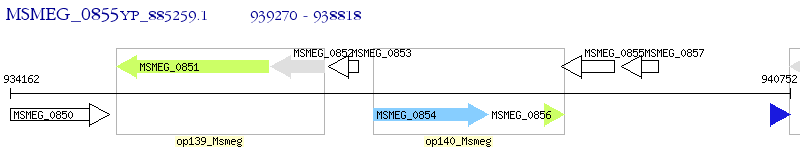
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0855 | - | - | 100% (150) | hypothetical protein MSMEG_0855 |
| M. smegmatis MC2 155 | MSMEG_6902 | - | 9e-06 | 44.23% (52) | hypothetical protein MSMEG_6902 |
| M. smegmatis MC2 155 | MSMEG_5505 | - | 2e-05 | 40.98% (61) | hypothetical protein MSMEG_5505 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0049c | - | 3e-07 | 36.08% (97) | hypothetical protein Mb0049c |
| M. gilvum PYR-GCK | Mflv_0159 | - | 3e-44 | 66.40% (125) | hypothetical protein Mflv_0159 |
| M. tuberculosis H37Rv | Rv0048c | - | 3e-07 | 36.08% (97) | hypothetical protein Rv0048c |
| M. leprae Br4923 | MLBr_00169 | - | 1e-05 | 35.06% (77) | hypothetical protein MLBr_00169 |
| M. abscessus ATCC 19977 | MAB_1068c | - | 4e-07 | 45.00% (60) | hypothetical protein MAB_1068c |
| M. marinum M | MMAR_0105 | - | 2e-11 | 40.00% (75) | hypothetical protein MMAR_0105 |
| M. avium 104 | MAV_1086 | - | 4e-06 | 39.13% (69) | hypothetical protein MAV_1086 |
| M. thermoresistible (build 8) | TH_1733 | - | 3e-07 | 36.11% (108) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4709 | - | 2e-06 | 42.03% (69) | hypothetical protein MUL_4709 |
| M. vanbaalenii PYR-1 | Mvan_0748 | - | 8e-44 | 59.42% (138) | hypothetical protein Mvan_0748 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0159|M.gilvum_PYR-GCK ------------MTSPTLRTGDREREATADVLGQALAQGYLDMTEYESRL
Mvan_0748|M.vanbaalenii_PYR-1 -----MSSIATPAATPSFRAGDRDRDMTADLLGQALAQGYLDMAEYESRL
MSMEG_0855|M.smegmatis_MC2_155 -----MANTVLPTVERSTRVGDPERERTADDLGRALAQGYLGMPEYEERL
MLBr_00169|M.leprae_Br4923 -MSNSAQRDAKGARDEPLRAADTDRIQIAQLLAYAAEQGRLELKDYEDRL
MAV_1086|M.avium_104 MMSNSTQRDAEDTRDEPSRAADTDRIQIAQLLAYAAEQGRLQLNDYEDRL
MUL_4709|M.ulcerans_Agy99 -MSNPARRDAQDTRDDSSLAADTDRIQIAQLLAYAAEQGRLQLNEYEDRL
TH_1733|M.thermoresistible__bu -----------------MRAADTDRIQMAHLLTDAAAQGRLRMDEYEERL
MAB_1068c|M.abscessus_ATCC_199 -MSNLPER-----VTASMRAADVDRMRVAQLLSDAAAQGRLELSEFEQRL
Mb0049c|M.bovis_AF2122/97 -MAKWLGAPLARGVSTATRAKDSDRQDACRILDDALRDGELSMEEHRERV
Rv0048c|M.tuberculosis_H37Rv -MAKWLGAPLARGVSTATRAKDSDRQDACRILDDALRDGELSMEEHRERV
MMAR_0105|M.marinum_M --------------------------------------------------
Mflv_0159|M.gilvum_PYR-GCK LAVYDAYTTPDLRQLTADLPVADLRRNDPRR-------------------
Mvan_0748|M.vanbaalenii_PYR-1 QTVYDARTTPDLRRLTSDLPVAELRRNDPRR-------------------
MSMEG_0855|M.smegmatis_MC2_155 QRVFGAQTTAQLRDLVADLPVAALRRNDPRR-------------------
MLBr_00169|M.leprae_Br4923 AKAYAATTYQELEQLRDDLPGSQVSARRGGNPNPAP------------ST
MAV_1086|M.avium_104 TRAYAAKTYEELDQLRADLPGAPISPRRGGKPNPAP------------ST
MUL_4709|M.ulcerans_Agy99 TRAYAATTYQELDQLSADLPGAAVSPRRGGNCNPAP------------ST
TH_1733|M.thermoresistible__bu TKAYAAKTYDELEQLSADLPGVVARRRSGGSVHPAP------------SS
MAB_1068c|M.abscessus_ATCC_199 AKVYSANTYGELDQLSADLPDVATSPVRGQGSKPAP------------ST
Mb0049c|M.bovis_AF2122/97 SAATKAVTLGDLQRLVADLQVESAPAQMPALKSRAKRTELGLLAAAFVAS
Rv0048c|M.tuberculosis_H37Rv SAATKAVTLGDLQRLVADLQVESAPAQMPALKSRAKRTELGLLAAAFVAS
MMAR_0105|M.marinum_M ------------------------MTDEQRR-------------------
Mflv_0159|M.gilvum_PYR-GCK --QAARTAAARRG-------------------------------------
Mvan_0748|M.vanbaalenii_PYR-1 --MAARRAAARRG-------------------------------------
MSMEG_0855|M.smegmatis_MC2_155 --REAQMRAARMS-------------------------------------
MLBr_00169|M.leprae_Br4923 LLLALMSGFERRGRWNVPRKLTT-------------------FSLWG--S
MAV_1086|M.avium_104 LLLALLSGFERRGRWNVPKKLTT-------------------FTLWG--S
MUL_4709|M.ulcerans_Agy99 LLLALLSGFERRGRWNVPKKLTT-------------------LTLWG--S
TH_1733|M.thermoresistible__bu LLLAIMGGFERRGRWNVPSRLTA-------------------IALFG--G
MAB_1068c|M.abscessus_ATCC_199 VLMAIMSGFERRGRWNVPGKLTS-------------------FALWG--G
Mb0049c|M.bovis_AF2122/97 VLLGVGIGWGVYGNTRSPLDFTSDPGAKPDGIAPVVLTPPRQLHSLGGLT
Rv0048c|M.tuberculosis_H37Rv VLLGVGIGWGVYGNTRSPLDFTSDPGAKPDGIAPVVLTPPRQLHSLGGLT
MMAR_0105|M.marinum_M --QALARIHAKRS-------------------------------------
.
Mflv_0159|M.gilvum_PYR-GCK ---------------------------------IHVHLAGYAAMVVIVLT
Mvan_0748|M.vanbaalenii_PYR-1 ---------------------------------VHVHLAGYLAMVVIVLT
MSMEG_0855|M.smegmatis_MC2_155 ---------------------------------VRLHLAAYVAGSVLMLG
MLBr_00169|M.leprae_Br4923 GVLDLR--------------YADFT-------STEVELHAYSVMGVQTIL
MAV_1086|M.avium_104 GVVDLR--------------YADFT-------STEVDIHAMSIMGVQSIL
MUL_4709|M.ulcerans_Agy99 GVLDLR--------------YADFT-------STEVEIHAYSIMGVQTIL
TH_1733|M.thermoresistible__bu GVIDLR--------------YADFT-------APEVVIRSYSICGGQTIL
MAB_1068c|M.abscessus_ATCC_199 GVLDLR--------------YADFT-------SNEVEIRSFSIMGGQTIL
Mb0049c|M.bovis_AF2122/97 GLLEQTRKRFGDTMGYRLVIYPEYASLDRVDPADDRRVLAYTYRGGWGDA
Rv0048c|M.tuberculosis_H37Rv GLLEQTRKRFGDTMGYRLVIYPEYASLDRVDPADDRRVLAYTYRGGWGDA
MMAR_0105|M.marinum_M ---------------------------------FWWHLGAYILGNAALVA
: .
Mflv_0159|M.gilvum_PYR-GCK VWLAVGLGSGGWYFWPVWPILGAGIGVLVHALPIRLAMPPCPGFTGHRSH
Mvan_0748|M.vanbaalenii_PYR-1 VWLAVGLTAGSWYFWPVWPILGAGVGVLAHALPIKHAFPACPGARRLASV
MSMEG_0855|M.smegmatis_MC2_155 IWLAVGLGGGGWYFWPVWPIMGWGIGVASHAIPIWAHGSMRP--ARIA--
MLBr_00169|M.leprae_Br4923 LPPEVNVEISGHGVMGSFDRQVRGQGTPGAPTVK-IRGFSLWGGVGIKRK
MAV_1086|M.avium_104 LPPEVNVEVHGRGVMGNFDRDVPGQGTPGAPTVH-IRGFSLWGGVGIRRK
MUL_4709|M.ulcerans_Agy99 LPPEINVEIDGRGIMGNFDREVAREGSRGAPNVK-IQGFSFWGGVGIKRK
TH_1733|M.thermoresistible__bu VPPEANVDLRGVGVMGSFDERVSGEGTPGAPCVR-IRGVAFLGNVKVRRK
MAB_1068c|M.abscessus_ATCC_199 LPPEVNVDVKGVAVMGGFDHSASGNGVQGAPHVT-VKGFSLWGGVNVKRK
Mb0049c|M.bovis_AF2122/97 TSSAKSIADVSVVDLSKFDAKTAVSIMRGAPETLGLKQSDVKSMYLIVEP
Rv0048c|M.tuberculosis_H37Rv TSSAKSIADVSVVDLSKFDAKTAVGIMRGAPETLGLKQSDVKSMYLIVEP
MMAR_0105|M.marinum_M IWF---FTSGG-YFWPIWPALGWGIGLVFHGLGVFLGMRPITEEQIQREI
. . :
Mflv_0159|M.gilvum_PYR-GCK H---------------------------------------
Mvan_0748|M.vanbaalenii_PYR-1 HSR-------------------------------------
MSMEG_0855|M.smegmatis_MC2_155 ----------------------------------------
MLBr_00169|M.leprae_Br4923 ARRPRR----------------------------------
MAV_1086|M.avium_104 ARRPRP----------------------------------
MUL_4709|M.ulcerans_Agy99 PRKARK----------------------------------
TH_1733|M.thermoresistible__bu RRKISSS---------------------------------
MAB_1068c|M.abscessus_ATCC_199 RRKRRESLEER-----------------------------
Mb0049c|M.bovis_AF2122/97 AKDPTTPAALSLSLYVSSDYGGGYLVFAGDGTIKHVSYPS
Rv0048c|M.tuberculosis_H37Rv VKDPTTPAALSLSLYVSSDYGGGYLVFAGDGTIKHVSYPS
MMAR_0105|M.marinum_M DRGHLS----------------------------------