For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSNSAQRDAKGARDEPLRAADTDRIQIAQLLAYAAEQGRLELKDYEDRLAKAYAATTYQELEQLRDDLPG SQVSARRGGNPNPAPSTLLLALMSGFERRGRWNVPRKLTTFSLWGSGVLDLRYADFTSTEVELHAYSVMG VQTILLPPEVNVEISGHGVMGSFDRQVRGQGTPGAPTVKIRGFSLWGGVGIKRKARRPRR
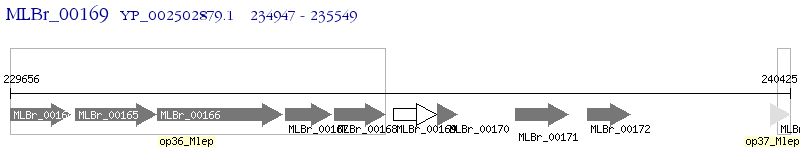
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00169 | - | - | 100% (200) | hypothetical protein MLBr_00169 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0991c | - | 2e-90 | 79.50% (200) | hypothetical protein Mb0991c |
| M. gilvum PYR-GCK | Mflv_1879 | - | 4e-68 | 65.78% (187) | hypothetical protein Mflv_1879 |
| M. tuberculosis H37Rv | Rv0966c | - | 2e-91 | 79.50% (200) | hypothetical protein Rv0966c |
| M. abscessus ATCC 19977 | MAB_1068c | - | 2e-66 | 64.48% (183) | hypothetical protein MAB_1068c |
| M. marinum M | MMAR_4539 | - | 2e-89 | 78.00% (200) | hypothetical protein MMAR_4539 |
| M. avium 104 | MAV_1086 | - | 3e-95 | 82.41% (199) | hypothetical protein MAV_1086 |
| M. smegmatis MC2 155 | MSMEG_5505 | - | 3e-61 | 60.41% (197) | hypothetical protein MSMEG_5505 |
| M. thermoresistible (build 8) | TH_1733 | - | 3e-63 | 62.98% (181) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4709 | - | 5e-89 | 77.50% (200) | hypothetical protein MUL_4709 |
| M. vanbaalenii PYR-1 | Mvan_4852 | - | 2e-66 | 64.52% (186) | hypothetical protein Mvan_4852 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1879|M.gilvum_PYR-GCK -MS------TPASRSGSMRAADTDRIQVAQLLTDAAASGKLGLTEYENRL
Mvan_4852|M.vanbaalenii_PYR-1 -MS------TPASRSGAMRAGDTDRIQVAQLLTDAAASGTLGLSEYESRL
TH_1733|M.thermoresistible__bu -----------------MRAADTDRIQMAHLLTDAAAQGRLRMDEYEERL
MSMEG_5505|M.smegmatis_MC2_155 -MS------TSAPQNGSMRAADTDRIQVAQQLTDAAASGRLPMDEYEDRL
MAB_1068c|M.abscessus_ATCC_199 -MSN-----LPERVTASMRAADVDRMRVAQLLSDAAAQGRLELSEFEQRL
MLBr_00169|M.leprae_Br4923 -MSNSAQRDAKGARDEPLRAADTDRIQIAQLLAYAAEQGRLELKDYEDRL
MAV_1086|M.avium_104 MMSNSTQRDAEDTRDEPSRAADTDRIQIAQLLAYAAEQGRLQLNDYEDRL
MMAR_4539|M.marinum_M -MSNPARRDAQDTRDDSSLAADTDRIQIAQLLAYAAEQGRLQLNEYEDRL
MUL_4709|M.ulcerans_Agy99 -MSNPARRDAQDTRDDSSLAADTDRIQIAQLLAYAAEQGRLQLNEYEDRL
Mb0991c|M.bovis_AF2122/97 -MSNSAQRDARNSRDESARASDTDRIQIAQLLAYAAEQGRLQLTDYEDRL
Rv0966c|M.tuberculosis_H37Rv -MSNSAQRDARNSRDESARASDTDRIQIAQLLAYAAEQGRLQLTDYEDRL
*.*.**:::*: *: ** .* * : ::*.**
Mflv_1879|M.gilvum_PYR-GCK NRAYAAQTYDELDRLSADLPGAVTRGR-SGPCRPAPSSMLLAILSGYERR
Mvan_4852|M.vanbaalenii_PYR-1 SRAYAAQTYDELDRLSADLPGAVTRGR-GGPCRPAPSTLLLAILTGYERR
TH_1733|M.thermoresistible__bu TKAYAAKTYDELEQLSADLPGVVARRRSGGSVHPAPSSLLLAIMGGFERR
MSMEG_5505|M.smegmatis_MC2_155 AKAYTAETRNELARLSSDLAGAATYLN--AGCRPAPSTTLLGLMSGFERR
MAB_1068c|M.abscessus_ATCC_199 AKVYSANTYGELDQLSADLPDVATSPVRGQGSKPAPSTVLMAIMSGFERR
MLBr_00169|M.leprae_Br4923 AKAYAATTYQELEQLRDDLPGSQVSARRGGNPNPAPSTLLLALMSGFERR
MAV_1086|M.avium_104 TRAYAAKTYEELDQLRADLPGAPISPRRGGKPNPAPSTLLLALLSGFERR
MMAR_4539|M.marinum_M TRAYAATTYQELDQLSADLPGAAVSPRRGGNCNPAPSTLLLALLSGFERR
MUL_4709|M.ulcerans_Agy99 TRAYAATTYQELDQLSADLPGAAVSPRRGGNCNPAPSTLLLALLSGFERR
Mb0991c|M.bovis_AF2122/97 ARAYAATTYQELDRLRADLPGAAIGPRRGGECNPAPSTLLLALLGGFERR
Rv0966c|M.tuberculosis_H37Rv ARAYAATTYQELDRLRADLPGAAIGPRRGGECNPAPSTLLLALLGGFERR
:.*:* * ** :* **.. .****: *:.:: *:***
Mflv_1879|M.gilvum_PYR-GCK GRWNVPRKLTTFALWGGGVVDLRYADFTSPEVDIRAYSVMGGQTILLPPE
Mvan_4852|M.vanbaalenii_PYR-1 GRWNVPRRLTTFALWGGGVVDLRYADFTSPEVDIRSYSIMGGQTILVPPE
TH_1733|M.thermoresistible__bu GRWNVPSRLTAIALFGGGVIDLRYADFTAPEVVIRSYSICGGQTILVPPE
MSMEG_5505|M.smegmatis_MC2_155 GRWNVPKKLTTFALFGGGVIDLRYADFTSADVDIRCYSIFGGQTILVPPE
MAB_1068c|M.abscessus_ATCC_199 GRWNVPGKLTSFALWGGGVLDLRYADFTSNEVEIRSFSIMGGQTILLPPE
MLBr_00169|M.leprae_Br4923 GRWNVPRKLTTFSLWGSGVLDLRYADFTSTEVELHAYSVMGVQTILLPPE
MAV_1086|M.avium_104 GRWNVPKKLTTFTLWGSGVVDLRYADFTSTEVDIHAMSIMGVQSILLPPE
MMAR_4539|M.marinum_M GRWNVPKKLTTLTLWGSGVLDLRYADFTSTEVEIHAYSIMGVQTILLPPE
MUL_4709|M.ulcerans_Agy99 GRWNVPKKLTTLTLWGSGVLDLRYADFTSTEVEIHAYSIMGVQTILLPPE
Mb0991c|M.bovis_AF2122/97 GRWNVPKKLTTFTLWGSGVLDLRYADFTSTEVDIRAYSIMGAQTILLPPE
Rv0966c|M.tuberculosis_H37Rv GRWNVPKKLTTFTLWGSGVLDLRYADFTSTEVDIRAYSIMGAQTILLPPE
****** :**:::*:*.**:********: :* ::. *: * *:**:***
Mflv_1879|M.gilvum_PYR-GCK VNVDLHGVAVMGSFDHSVDGEGSPGAPLVRITGFSVWGSVSVKRKRRKPA
Mvan_4852|M.vanbaalenii_PYR-1 VNVDSRGVAVMGSFDHGVDGEGSAGAPLVRIRGFALWGSVGVKRKRRKAP
TH_1733|M.thermoresistible__bu ANVDLRGVGVMGSFDERVSGEGTPGAPCVRIRGVAFLGNVKVRRKRRKIS
MSMEG_5505|M.smegmatis_MC2_155 VNVDCHGIGIMGNFDRHVHGEGVPGAPRVHIRGFSVGGTVSVKRKHRRKP
MAB_1068c|M.abscessus_ATCC_199 VNVDVKGVAVMGGFDHSASGNGVQGAPHVTVKGFSLWGGVNVKRKRRKRR
MLBr_00169|M.leprae_Br4923 VNVEISGHGVMGSFDRQVRGQGTPGAPTVKIRGFSLWGGVGIKRKARRPR
MAV_1086|M.avium_104 VNVEVHGRGVMGNFDRDVPGQGTPGAPTVHIRGFSLWGGVGIRRKARRPR
MMAR_4539|M.marinum_M INVEIDGRGIMGNFDREVAGDGSRGAPKVKIQGFSFWGGVGIKRKPRKAR
MUL_4709|M.ulcerans_Agy99 INVEIDGRGIMGNFDREVAREGSRGAPNVKIQGFSFWGGVGIKRKPRKAR
Mb0991c|M.bovis_AF2122/97 VNVEIHGHRVMGGFDRKVVGEGTRGAPTVRIRGFSLGGDVGIKRKPRKPR
Rv0966c|M.tuberculosis_H37Rv VNVEIHGHRVMGGFDRKVVGEGTRGVPTVRIRGFSLWGDVGIKRKPRKPR
**: * :**.**. . :* *.* * : *.:. * * ::** *:
Mflv_1879|M.gilvum_PYR-GCK A-----
Mvan_4852|M.vanbaalenii_PYR-1 ADTAEG
TH_1733|M.thermoresistible__bu SS----
MSMEG_5505|M.smegmatis_MC2_155 HEN---
MAB_1068c|M.abscessus_ATCC_199 ESLEER
MLBr_00169|M.leprae_Br4923 R-----
MAV_1086|M.avium_104 P-----
MMAR_4539|M.marinum_M K-----
MUL_4709|M.ulcerans_Agy99 K-----
Mb0991c|M.bovis_AF2122/97 K-----
Rv0966c|M.tuberculosis_H37Rv K-----