For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSIATPAATPSFRAGDRDRDMTADLLGQALAQGYLDMAEYESRLQTVYDARTTPDLRRLTSDLPVAELRR NDPRRMAARRAAARRGVHVHLAGYLAMVVIVLTVWLAVGLTAGSWYFWPVWPILGAGVGVLAHALPIKHA FPACPGARRLASVHSR*
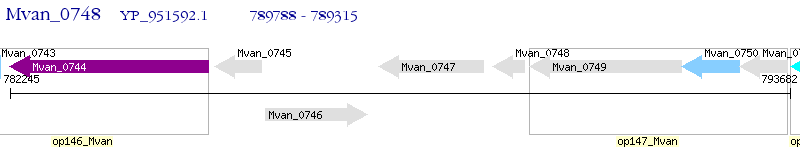
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0748 | - | - | 100% (157) | hypothetical protein Mvan_0748 |
| M. vanbaalenii PYR-1 | Mvan_4852 | - | 2e-10 | 48.05% (77) | hypothetical protein Mvan_4852 |
| M. vanbaalenii PYR-1 | Mvan_6038 | - | 2e-06 | 35.00% (100) | hypothetical protein Mvan_6038 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0991c | - | 1e-07 | 46.15% (65) | hypothetical protein Mb0991c |
| M. gilvum PYR-GCK | Mflv_0159 | - | 3e-63 | 79.71% (138) | hypothetical protein Mflv_0159 |
| M. tuberculosis H37Rv | Rv0966c | - | 1e-07 | 46.15% (65) | hypothetical protein Rv0966c |
| M. leprae Br4923 | MLBr_00169 | - | 3e-07 | 40.00% (65) | hypothetical protein MLBr_00169 |
| M. abscessus ATCC 19977 | MAB_1068c | - | 6e-09 | 50.00% (56) | hypothetical protein MAB_1068c |
| M. marinum M | MMAR_0105 | - | 4e-08 | 35.82% (67) | hypothetical protein MMAR_0105 |
| M. avium 104 | MAV_1086 | - | 4e-07 | 44.44% (63) | hypothetical protein MAV_1086 |
| M. smegmatis MC2 155 | MSMEG_0855 | - | 9e-44 | 59.42% (138) | hypothetical protein MSMEG_0855 |
| M. thermoresistible (build 8) | TH_1733 | - | 2e-08 | 50.00% (58) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4709 | - | 1e-06 | 45.16% (62) | hypothetical protein MUL_4709 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0991c|M.bovis_AF2122/97 -MSNSAQRDARNSRDESARASDTDRIQIAQLLAYAAEQGRLQLTDYEDRL
Rv0966c|M.tuberculosis_H37Rv -MSNSAQRDARNSRDESARASDTDRIQIAQLLAYAAEQGRLQLTDYEDRL
MUL_4709|M.ulcerans_Agy99 -MSNPARRDAQDTRDDSSLAADTDRIQIAQLLAYAAEQGRLQLNEYEDRL
MLBr_00169|M.leprae_Br4923 -MSNSAQRDAKGARDEPLRAADTDRIQIAQLLAYAAEQGRLELKDYEDRL
MAV_1086|M.avium_104 MMSNSTQRDAEDTRDEPSRAADTDRIQIAQLLAYAAEQGRLQLNDYEDRL
TH_1733|M.thermoresistible__bu -----------------MRAADTDRIQMAHLLTDAAAQGRLRMDEYEERL
MAB_1068c|M.abscessus_ATCC_199 -MSNLPER-----VTASMRAADVDRMRVAQLLSDAAAQGRLELSEFEQRL
Mvan_0748|M.vanbaalenii_PYR-1 -----MSSIATPAATPSFRAGDRDRDMTADLLGQALAQGYLDMAEYESRL
Mflv_0159|M.gilvum_PYR-GCK ------------MTSPTLRTGDREREATADVLGQALAQGYLDMTEYESRL
MSMEG_0855|M.smegmatis_MC2_155 -----MANTVLPTVERSTRVGDPERERTADDLGRALAQGYLGMPEYEERL
MMAR_0105|M.marinum_M --------------------------------------------------
Mb0991c|M.bovis_AF2122/97 ARAYAATTYQELDRLRADLPGAAIGPRRGGECNPAPSTLLLALLGGFERR
Rv0966c|M.tuberculosis_H37Rv ARAYAATTYQELDRLRADLPGAAIGPRRGGECNPAPSTLLLALLGGFERR
MUL_4709|M.ulcerans_Agy99 TRAYAATTYQELDQLSADLPGAAVSPRRGGNCNPAPSTLLLALLSGFERR
MLBr_00169|M.leprae_Br4923 AKAYAATTYQELEQLRDDLPGSQVSARRGGNPNPAPSTLLLALMSGFERR
MAV_1086|M.avium_104 TRAYAAKTYEELDQLRADLPGAPISPRRGGKPNPAPSTLLLALLSGFERR
TH_1733|M.thermoresistible__bu TKAYAAKTYDELEQLSADLPGVVARRRSGGSVHPAPSSLLLAIMGGFERR
MAB_1068c|M.abscessus_ATCC_199 AKVYSANTYGELDQLSADLPDVATSPVRGQGSKPAPSTVLMAIMSGFERR
Mvan_0748|M.vanbaalenii_PYR-1 QTVYDARTTPDLRRLTSDLPVAELRR------------------------
Mflv_0159|M.gilvum_PYR-GCK LAVYDAYTTPDLRQLTADLPVADLRR------------------------
MSMEG_0855|M.smegmatis_MC2_155 QRVFGAQTTAQLRDLVADLPVAALRR------------------------
MMAR_0105|M.marinum_M -----------------------MT-------------------------
Mb0991c|M.bovis_AF2122/97 GRWNVPKKLTTFTLWGSGVLDLRYADFTSTEVDIRAYSIMGAQTILLPPE
Rv0966c|M.tuberculosis_H37Rv GRWNVPKKLTTFTLWGSGVLDLRYADFTSTEVDIRAYSIMGAQTILLPPE
MUL_4709|M.ulcerans_Agy99 GRWNVPKKLTTLTLWGSGVLDLRYADFTSTEVEIHAYSIMGVQTILLPPE
MLBr_00169|M.leprae_Br4923 GRWNVPRKLTTFSLWGSGVLDLRYADFTSTEVELHAYSVMGVQTILLPPE
MAV_1086|M.avium_104 GRWNVPKKLTTFTLWGSGVVDLRYADFTSTEVDIHAMSIMGVQSILLPPE
TH_1733|M.thermoresistible__bu GRWNVPSRLTAIALFGGGVIDLRYADFTAPEVVIRSYSICGGQTILVPPE
MAB_1068c|M.abscessus_ATCC_199 GRWNVPGKLTSFALWGGGVLDLRYADFTSNEVEIRSFSIMGGQTILLPPE
Mvan_0748|M.vanbaalenii_PYR-1 ---NDPRRMAARRAAARRGVHVHLAGYLAMVVIVLTVWLA----------
Mflv_0159|M.gilvum_PYR-GCK ---NDPRRQAARTAAARRGIHVHLAGYAAMVVIVLTVWLA----------
MSMEG_0855|M.smegmatis_MC2_155 ---NDPRRREAQMRAARMSVRLHLAAYVAGSVLMLGIWLA----------
MMAR_0105|M.marinum_M ---DEQRRQALARIHAKRSFWWHLGAYILGNAALVAIWFF----------
: : . . : . : . : .
Mb0991c|M.bovis_AF2122/97 VNVEIHGHRVMGGFDRKVVGEGTRGAPTVRIRGFSLGGDVGIKRKPRKPR
Rv0966c|M.tuberculosis_H37Rv VNVEIHGHRVMGGFDRKVVGEGTRGVPTVRIRGFSLWGDVGIKRKPRKPR
MUL_4709|M.ulcerans_Agy99 INVEIDGRGIMGNFDREVAREGSRGAPNVKIQGFSFWGGVGIKRKPRKAR
MLBr_00169|M.leprae_Br4923 VNVEISGHGVMGSFDRQVRGQGTPGAPTVKIRGFSLWGGVGIKRKARRPR
MAV_1086|M.avium_104 VNVEVHGRGVMGNFDRDVPGQGTPGAPTVHIRGFSLWGGVGIRRKARRPR
TH_1733|M.thermoresistible__bu ANVDLRGVGVMGSFDERVSGEGTPGAPCVRIRGVAFLGNVKVRRKRRKIS
MAB_1068c|M.abscessus_ATCC_199 VNVDVKGVAVMGGFDHSASGNGVQGAPHVTVKGFSLWGGVNVKRKRRKRR
Mvan_0748|M.vanbaalenii_PYR-1 VGLTAGSWYFWPVWPILGAGVG------VLAHALPIKHAFPACPGARRLA
Mflv_0159|M.gilvum_PYR-GCK VGLGSGGWYFWPVWPILGAGIG------VLVHALPIRLAMPPCPGFTGHR
MSMEG_0855|M.smegmatis_MC2_155 VGLGGGGWYFWPVWPIMGWGIG------VASHAIPIWAHGSMRP--ARIA
MMAR_0105|M.marinum_M ---TSGG-YFWPIWPALGWGIG------LVFHGLGVFLGMRPITEEQIQR
. . : * : :.. .
Mb0991c|M.bovis_AF2122/97 K-------
Rv0966c|M.tuberculosis_H37Rv K-------
MUL_4709|M.ulcerans_Agy99 K-------
MLBr_00169|M.leprae_Br4923 R-------
MAV_1086|M.avium_104 P-------
TH_1733|M.thermoresistible__bu SS------
MAB_1068c|M.abscessus_ATCC_199 ESLEER--
Mvan_0748|M.vanbaalenii_PYR-1 SVHSR---
Mflv_0159|M.gilvum_PYR-GCK SHH-----
MSMEG_0855|M.smegmatis_MC2_155 --------
MMAR_0105|M.marinum_M EIDRGHLS