For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQVPGPFEYERATSVDHAVGLLDRLGEDARIVAGGHSLLPMMKLRIANPEYLVDINDLAVELGYVITDPT LVRIGAMARHRQVLESDPLAAVCPIFRDAERVIADPVVRNRGTLGGSLCQADPAEDLTTVCTILGAVCLA RGPGGEREIGIDDFLVGPYETALAHNEMLVEVRIPVRHRTSSAYAKVERRVGDWAVTAAGAQVTLDGDSI VAARVGLTAVNPDPDALRALADDLIGKPATEETFAAAGELAVQACEPVTDTRGSADYKRHLARELTIRTM RTAVERVRTAPAPEGN
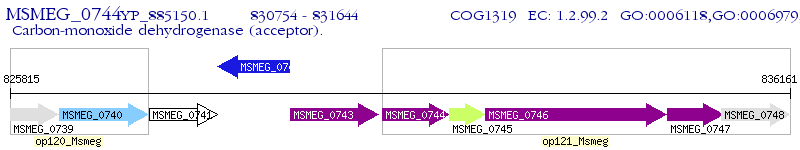
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0744 | - | - | 100% (296) | carbon monoxide dehydrogenase medium chain |
| M. smegmatis MC2 155 | MSMEG_5882 | - | 1e-35 | 36.95% (295) | carbon monoxide dehydrogenase medium chain |
| M. smegmatis MC2 155 | MSMEG_2463 | - | 2e-30 | 31.94% (288) | nicotine dehydrogenase chain A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0382c | - | 1e-128 | 80.14% (282) | carbon monoxyde dehydrogenase medium subunit |
| M. gilvum PYR-GCK | Mflv_1571 | - | 6e-35 | 33.68% (288) | carbon-monoxide dehydrogenase (acceptor) |
| M. tuberculosis H37Rv | Rv0375c | - | 1e-128 | 80.14% (282) | carbon monoxyde dehydrogenase medium subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2926c | - | 8e-07 | 27.24% (268) | putative oxidoreductase (molybdopterin dehydrogenase) |
| M. marinum M | MMAR_0655 | coxM | 1e-141 | 84.12% (296) | carbon monoxyde dehydrogenase (medium chain), CoxM |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0174 | - | 1e-130 | 78.12% (288) | PROBABLE CARBON MONOXYDE DEHYDROGENASE (MEDIUM CHAIN) |
| M. ulcerans Agy99 | MUL_0587 | coxM_2 | 1e-36 | 33.56% (292) | aerobic-type carbon monoxide dehydrogenase, CoxM_2 |
| M. vanbaalenii PYR-1 | Mvan_5185 | - | 4e-34 | 34.16% (281) | carbon-monoxide dehydrogenase (acceptor) |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1571|M.gilvum_PYR-GCK -MKAAPFAYHRPDTVADAVSLLGEYGDDAKILAGGQSLVPMLAMRLTHFE
Mvan_5185|M.vanbaalenii_PYR-1 -MKAAPFAYHRPDTVADAVSMLGEFGDDAKILAGGQSLVPMLAMRLTHFE
MUL_0587|M.ulcerans_Agy99 -MKPASFDYHRPDTAEEAVDLLAELGEDAKILPGGQSLVPMLSMRLAFFD
MSMEG_0744|M.smegmatis_MC2_155 MQVPGPFEYERATSVDHAVGLLDRLGEDARIVAGGHSLLPMMKLRIANPE
MMAR_0655|M.marinum_M MQVPGPFEYERATSVDHAIGLLDRLGEGARLVAGGHSLLPMMKLRIANPE
Mb0382c|M.bovis_AF2122/97 --------------MDHAIGLLDRLGEGARVVAGGHSLLPMMKLRIANPE
Rv0375c|M.tuberculosis_H37Rv --------------MDHAIGLLDRLGEGARVVAGGHSLLPMMKLRIANPE
TH_0174|M.thermoresistible__bu MQVPGPFEYERATSVDHAIGLLDRLGDGAMIVAGGHSLLPMMKLRIANPE
MAB_2926c|M.abscessus_ATCC_199 ----MDLYSIDSVLIPRDRADLSGLGTQDGVLAGGTWLMSQPQPGLR---
* * ::.** *:. :
Mflv_1571|M.gilvum_PYR-GCK NLIDISRIEELRNIELRNG--ADSSVRIGAGTPHALVGMDDEVAESVPLL
Mvan_5185|M.vanbaalenii_PYR-1 NLIDISRVDELRNIEPRG-----DAVRIGAGTPHALVGMDDEVAESVPLL
MUL_0587|M.ulcerans_Agy99 HLIDISRLGELQGIRLRD-----DQLWIGAGTPDSKVGTNSQVHQSIPLL
MSMEG_0744|M.smegmatis_MC2_155 YLVDINDLAVELGYVIT----DPTLVRIGAMARHRQVLESDPLAAVCPIF
MMAR_0655|M.marinum_M YLVDINDLAPELGYVIT----DPTLVRIGAMARHREILESEALAAVCPIF
Mb0382c|M.bovis_AF2122/97 YLVDINDLAPELGYVVVGGINNPNLVRLGAMTRHREILDSDALAAVCPIF
Rv0375c|M.tuberculosis_H37Rv YLVDINDLAPELGYVVVGGINNPNLVRLGAMTRHREILDSDALAAVCPIF
TH_0174|M.thermoresistible__bu YLVDINDLAGELGYIVV----NADAVRIGAMTRHREALESAELAQCCPIF
MAB_2926c|M.abscessus_ATCC_199 RLVDLTGMRWPDLVVDAGG---LEVAATCSIERLINASYPPAWQATALFG
*:*:. : : :
Mflv_1571|M.gilvum_PYR-GCK TLATPHIGHFQIRTRGTLGGAIAHADPAAEYAAVALALDAEIEATSGRGT
Mvan_5185|M.vanbaalenii_PYR-1 TLSTPHIGHFQIRTRGTLGGAIAHADPAAEYAAVALALDAEMEATSARGT
MUL_0587|M.ulcerans_Agy99 ARATPYIGHFQIRNRGTLGGSIAHADAAGEYPTVALTLDAVMEVLSPRGH
MSMEG_0744|M.smegmatis_MC2_155 RDAERVIADPVVRNRGTLGGSLCQADPAEDLTTVCTILGAVCLARGPGGE
MMAR_0655|M.marinum_M RDAERVIADPVVRNRGTLGGSLCQADPAEDLSTVCTVLDAVCLARGSSGE
Mb0382c|M.bovis_AF2122/97 RDAERVIADPVVRNRGTLGGSLCQADPAEDLSTVCTVLDAVCLAKGPSGE
Rv0375c|M.tuberculosis_H37Rv RDAERVIADPVVRNRGTLGGSLCQADPAEDLSTVCTVLDAVCLAKGPSGE
TH_0174|M.thermoresistible__bu ADAERVIADPVVRNRGTVGGSLCQADPAEDLSTVCTVLDAVCVVRGPAGR
MAB_2926c|M.abscessus_ATCC_199 QCARALLASFKIWRTATVGGNICLGLPAGAMISLCAALDAELHIWRHDGT
: :. : .*:** :. . .* ::. *.* *
Mflv_1571|M.gilvum_PYR-GCK RR-IPAAEFFTGLWETALASDEILTAVSFPVWSGRCGFAVQEFARRHGDF
Mvan_5185|M.vanbaalenii_PYR-1 RR-IPAAEFFTGLWETALAADEILTAVSFPVWTGHCGFAVQEFARRHGDF
MUL_0587|M.ulcerans_Agy99 RK-IPAAEFFTGLWETAMEPDEVLTGVRFPTWTGKSGFAMREFSRRHGDF
MSMEG_0744|M.smegmatis_MC2_155 RE-IGIDDFLVGPYETALAHNEMLVEVRIPVRH-RTSSAYAKVERRVGDW
MMAR_0655|M.marinum_M RE-IAIDEFQAGPYETTLAPSEMLVEVRIPVRH-NTSSAYAKVERRVGDW
Mb0382c|M.bovis_AF2122/97 RE-IAIDDFLVGPYETALAHNEVLIEVRIPLRH-NTSSAYAKVERRVGDW
Rv0375c|M.tuberculosis_H37Rv RE-IAIDDFLVGPYETALAHNEVLIEVRIPLRH-NTSSAYAKVERRVGDW
TH_0174|M.thermoresistible__bu RE-IGIDDFLVGPYETALAHNEILLEVKIPLRP-NSSNAYAKVERRVGDW
MAB_2926c|M.abscessus_ATCC_199 DERCTITEFVTGINETVLHPGDVLRAITFPSSALSQPTALRKIAYSP--L
. :* .* **.: .::* : :* * :.
Mflv_1571|M.gilvum_PYR-GCK AIAGATVAVALDEDDTVTRCGIGMLGMGSTPERGTAAEAAVLGRPVADIT
Mvan_5185|M.vanbaalenii_PYR-1 AIAGATLAVELDGRDRVARCGIGLLGMGSTPERGTPAEQAVVGRPVTDIT
MUL_0587|M.ulcerans_Agy99 AIAGAAVAVQLDDEHRVSRCAIGLLGLGPTPRRATAAEAAVLGKPIDELV
MSMEG_0744|M.smegmatis_MC2_155 AVTAAGAQVTLDG-DSIVAARVGLTAVNPDPDALRALADDLIGKPATEET
MMAR_0655|M.marinum_M AVTAAGAAVTLNN-DTITAARVGLTAVNPDPAALAELAAALVGRPANEDT
Mb0382c|M.bovis_AF2122/97 AITAAGAAVTLDG-QTILAARVGLTAVNPDPVALAELSAGLVGQPATEEV
Rv0375c|M.tuberculosis_H37Rv AITAAGAAVTLDG-QTILAARVGLTAVNPDPVALAELSAGLVGQPATEEV
TH_0174|M.thermoresistible__bu AVTAAGAAVTLDG-QTVTAARIGLTAVNPDAAALAEISRRLTGRPATEET
MAB_2926c|M.abscessus_ATCC_199 GRSGALVTGRLDG----ADIVLGITAATRRP---------IVLRVPAEAR
. :.* *: :*: . . : : :
Mflv_1571|M.gilvum_PYR-GCK AEELGRLAVSTLHDIPADLQGSAAYRSRVCATMVARAWSEATTAARHTTE
Mvan_5185|M.vanbaalenii_PYR-1 AEEVGRLAVSELHDVPADLQGRPAYRSKVGATMVARAWSQATTAALETRE
MUL_0587|M.ulcerans_Agy99 PEQIGQSAMSGLDDIASDLQGPASYRAQVGAVMTTRAWSRAVQEAN--AE
MSMEG_0744|M.smegmatis_MC2_155 FAAAGELAVQACEPVT-DTRGSADYKRHLARELTIRTMRTAVERVRTAPA
MMAR_0655|M.marinum_M FAEAGRRAGQACEPVT-DIRGTADYKRHLAAELTIRTLRSAVGRVRNQPA
Mb0382c|M.bovis_AF2122/97 FAEAGRRAAQACTPVT-DVRGTAEYKRHLAGELTVRTLRTAAGRVLGAPA
Rv0375c|M.tuberculosis_H37Rv FAEAGRRAAQACTPVT-DVRGTAEYKRHLAGELTVRTLRTAAGRVLGAPA
TH_0174|M.thermoresistible__bu FAEAGRLAAQACEPVT-DVRGTADYKRHLAAELTTRTLRTAVQRIQQRHG
MAB_2926c|M.abscessus_ATCC_199 RAREAVAAIPGVEFHT-DAHGTAEWRGAVTTQLAGEVAAELTGALS----
. * . * :* . :: : :. .. .
Mflv_1571|M.gilvum_PYR-GCK ALHA--
Mvan_5185|M.vanbaalenii_PYR-1 ALNA--
MUL_0587|M.ulcerans_Agy99 TAHA--
MSMEG_0744|M.smegmatis_MC2_155 PEGN--
MMAR_0655|M.marinum_M PEGN--
Mb0382c|M.bovis_AF2122/97 APEA--
Rv0375c|M.tuberculosis_H37Rv APEA--
TH_0174|M.thermoresistible__bu GAQPCR
MAB_2926c|M.abscessus_ATCC_199 ------