For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLREPRAIGQPVIRKESRAKVTGAAPYAFEHIVDNALYLHPVQATIARGRVVAMDVAAARALDGVRGILT VFDAPELADTSDGEFAILQDDRVHFRGQIIGGVLAESAETAREAAALVRVEYLQEPHDTELRADHPGLYT PDEVNGGEEPDTDDGDVEAALAQAAVTVDETYETPIEHNNPMEPHACIAQWNLGDSGKPAVVLYDSTQGV HAVRKALAPIFGLEPEQIRVIAPHVGGGFGSKGAPHAHNVLAILAAQRSNGRPVKLALTRQQMFSLVGYR TPTIQRIRLGADHDGRITALVHDVVEQTSAVKEFAEQTAITSRKMYSAASRRTTHKLAALDVAIPFWMRA PGECPGTYAAEVAMDELAVACGLDPIELRVRNDPEVDPETGKPWSGRHLVDCLRLGADRFGWAARDPRPA ARRDGEWLIGTGVAAATYPGKAMPGNSARITYAAEGRYTVQIGAADIGTGTWTTLSQIAADALACDLAAV DLQIGDTDLPEASVAGGSSGITSWGSAIVAAARQFRADHGDPPAVGATTVAQSPENPDADGVTMQSFGAH FVEAHVNADTGEIRVPRMLGVFSVGRVINARTLRSQLIGGMTMGLSMALHEESVRDPRFGHVVTQDLATY HISAHADVADIDAIWLDEADEYLNPMGSRGAGEIGIVGSASAVVNAVYHATGVRVRNLPVTVDKVLAGLP KLV*
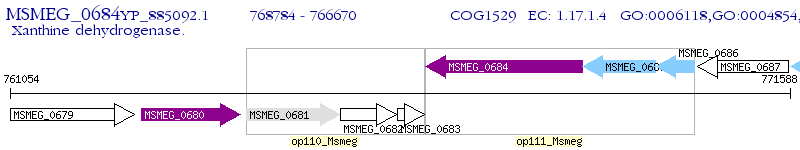
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0684 | - | - | 100% (704) | aldehyde oxidase and xanthine dehydrogenase, molybdopterin binding |
| M. smegmatis MC2 155 | MSMEG_0870 | - | e-115 | 36.94% (739) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_1291 | - | 2e-62 | 28.49% (744) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0380c | - | 2e-44 | 26.58% (775) | carbon monoxyde dehydrogenase large subunit |
| M. gilvum PYR-GCK | Mflv_0284 | - | 0.0 | 78.03% (701) | aldehyde oxidase and xanthine dehydrogenase, molybdopterin |
| M. tuberculosis H37Rv | Rv0373c | - | 4e-44 | 26.58% (775) | carbon monoxyde dehydrogenase large subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2925c | - | 1e-56 | 29.05% (747) | xanthine dehydrogenase family protein |
| M. marinum M | MMAR_0080 | coxL_2 | 1e-113 | 37.41% (703) | carbon monoxyde dehydrogenase (large chain), CoxL_2 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0176 | - | 2e-44 | 26.88% (770) | PROBABLE CARBON MONOXYDE DEHYDROGENASE (LARGE CHAIN) |
| M. ulcerans Agy99 | MUL_4580 | coxL_1 | 1e-106 | 34.64% (739) | carbon monoxyde dehydrogenase (large chain), CoxL_1 |
| M. vanbaalenii PYR-1 | Mvan_2075 | - | 1e-115 | 37.15% (724) | aldehyde oxidase and xanthine dehydrogenase, molybdopterin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0684|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0284|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4580|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2075|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0080|M.marinum_M --------------------------------------------------
Mb0380c|M.bovis_AF2122/97 --------------------------------------------------
Rv0373c|M.tuberculosis_H37Rv --------------------------------------------------
TH_0176|M.thermoresistible__bu --------------------------------------------------
MAB_2925c|M.abscessus_ATCC_199 MKVNGKLIDASPRPGQCLRTFLREHGHVEVKKGCDAGDCGACTVLVDGAS
MSMEG_0684|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0284|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4580|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2075|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0080|M.marinum_M --------------------------------------------------
Mb0380c|M.bovis_AF2122/97 --------------------------------------------------
Rv0373c|M.tuberculosis_H37Rv --------------------------------------------------
TH_0176|M.thermoresistible__bu --------------------------------------------------
MAB_2925c|M.abscessus_ATCC_199 QHSCIVPAHRVTDAEIHTVRSLAGEHDLHPMQRGFIDHAGFQCGFCTAGM
MSMEG_0684|M.smegmatis_MC2_155 ---------------------------------MTLREPRAIGQPVIRKE
Mflv_0284|M.gilvum_PYR-GCK ---------------------------------MTMLEPRAIGAPTARRD
MUL_4580|M.ulcerans_Agy99 ------------------------------MTHAFSAAGGISGQPINRVD
Mvan_2075|M.vanbaalenii_PYR-1 ----------------------------------MVAS---VGLPVNRVE
MMAR_0080|M.marinum_M ---------------------------------MTTAPATRGAMRFARVE
Mb0380c|M.bovis_AF2122/97 ------------------MTTIESRPPS-PEDLADNAQQPCGHGRMMRKE
Rv0373c|M.tuberculosis_H37Rv ------------------MTTIESRPPS-PEDLADNAQQPCGHGRMMRKE
TH_0176|M.thermoresistible__bu ------------------MTTLESNPAARPDDFENNDQKPCGHGRMLRKE
MAB_2925c|M.abscessus_ATCC_199 VLTAATLTGDQTEDLGNALKGNLCRCTGYRPIRDAVCGVVPPRGSVSAPA
MSMEG_0684|M.smegmatis_MC2_155 SRAKVTGAAPYAFEHIVDNALYLHPVQATIARGRVVAMDVAAARA----L
Mflv_0284|M.gilvum_PYR-GCK GQAKVTGAAAYAYEQQVDSPAYLHPVQATIARGRVIAMDTNLARA----V
MUL_4580|M.ulcerans_Agy99 GPLKVTGKARYAADHPIPGLVYAALVCSTVARGAVDHIATGAAMR----Q
Mvan_2075|M.vanbaalenii_PYR-1 GVDKVTGKARYSADTQVAGVAYAALVQAQIPHGRVTAESVDAAAATASAA
MMAR_0080|M.marinum_M AYSKITGTARYAADHRPEGLLYAVLVGAPVAAGRLKAVDADAALG----S
Mb0380c|M.bovis_AF2122/97 DPRFIRGRGTYVDDVALPGMLHLAILRSPYAHARIVRIDVTAAQA----H
Rv0373c|M.tuberculosis_H37Rv DPRFIRGRGTYVDDVALPGMLHLAILRSPYAHARIVRIDVTAAQA----H
TH_0176|M.thermoresistible__bu DPRFIRGRGRYVDDVQLPGMLHLAILRSPYAHARIAGIDTSAAQA----H
MAB_2925c|M.abscessus_ATCC_199 AARVVTGTEPYTMDELPAGVGHVAVLGSPHPSARITRIDTDRARL----L
: * * : . : : : . . : . *
MSMEG_0684|M.smegmatis_MC2_155 DGVRGILTVFDAPELADTSDG--------EFAILQDDRVHFRGQIIGGVL
Mflv_0284|M.gilvum_PYR-GCK DGVLDILTVSDAPKLADASDG--------ELAILQDATVHFRGQLIGGVV
MUL_4580|M.ulcerans_Agy99 PGVLAVLTDFTG-----------------VRLPFGTGQVFFFGQPIAVVV
Mvan_2075|M.vanbaalenii_PYR-1 PGVLYVLTPTNCPPLQVLPKELTWDLPLERRPPLSDLTVQHVGQHMALVV
MMAR_0080|M.marinum_M PGVVAVLIADDLPDFPLLSPP-----SAVRRMPLMNNEIHYEGQAVAIVI
Mb0380c|M.bovis_AF2122/97 PKVKAVVTGADLAAKGLAWMP---TLANDVQAVLATDKTRFQGQEVASVV
Rv0373c|M.tuberculosis_H37Rv PKVKAVVTGADLAAKGLAWMP---TLANDVQAVLATDKTRFQGQEVAFVV
TH_0176|M.thermoresistible__bu PKVKAVVTGADLAEKGLAWMP---TLSNDVQAVLATDKVRFQGQEVAFVV
MAB_2925c|M.abscessus_ATCC_199 PGVRAVLCYRDDPGVPFSTARHHERTDDPDDTLVFDRVLRFAGQRVAAVV
* :: . . ** :. *:
MSMEG_0684|M.smegmatis_MC2_155 AESAETAREAAALVRVEYLQEPHDTELRADHPGLYTPDEVNGGEEPDTDD
Mflv_0284|M.gilvum_PYR-GCK AETAEIAREAAALVHVEYQEKEHDAALRADHPGLYTPESVNPSYPPDSDQ
MUL_4580|M.ulcerans_Agy99 ATTHEAAAHAVSLVDVSYSTWPQLTDIDAPQAVGYPGRLIADYVRG---T
Mvan_2075|M.vanbaalenii_PYR-1 ADSPENAVHAASLMDFDYIAAPAHLDATEVVSGGGAADDVDELIRHGAYR
MMAR_0080|M.marinum_M AESIEAAEAARALVVVDCERTNAVVLGSGTREP-ADGEYSDPFDKG---D
Mb0380c|M.bovis_AF2122/97 AEDRYSARDACELVDVDYEPRDPVVDARTALDPSAPVIRTDLEGKSDNHI
Rv0373c|M.tuberculosis_H37Rv AEDRYSARDACELVDVDYEPRDPVVDARTALDPSAPVIRTDLEGKSDNHI
TH_0176|M.thermoresistible__bu AEDHYSARDALELIDVDYDPLDPVVDVRRALEPDAPVIRTDLDGKTDNHC
MAB_2925c|M.abscessus_ATCC_199 ADTPAIAHRALEHIDVEYEVLPAVFDPEMAQLPGAHLVHGDKGLGARIAD
* * * : ..
MSMEG_0684|M.smegmatis_MC2_155 GDVEAALA----------------QAAVTVDETYETPIEHNNPMEPHACI
Mflv_0284|M.gilvum_PYR-GCK GDVESAFP----------------AAEFTVDQTYRTPTEHNNPMEPHACV
MUL_4580|M.ulcerans_Agy99 PDQVMIR-----------------AAGIS-DLNFSIARNNHNAVELPATI
Mvan_2075|M.vanbaalenii_PYR-1 PDHFVKLTDEKLQ---DFRGPRGEPPGIKVDQTYRTAVNAHYPIELSATV
MMAR_0080|M.marinum_M FDTAWSN------------------AAIRVERTYLQPPRHHHAMETSGTV
Mb0380c|M.bovis_AF2122/97 FDWETGDAAAT--------EAVFAKADVVVQQEIVYPRVHPAPMETCGAV
Rv0373c|M.tuberculosis_H37Rv FDWETGDAAAT--------EAVFAKADVVVQQEIVYPRVHPAPMETCGAV
TH_0176|M.thermoresistible__bu FDWETGDPAAT--------EAAFAKADVVVEQEIVYPRVHPAPMETCGAV
MAB_2925c|M.abscessus_ATCC_199 PARNLVAEVHGEVGDVAAALARARESGAVVTGSWQTARVQHVHLETHGSV
. . :* . :
MSMEG_0684|M.smegmatis_MC2_155 AQWNLGDSGKPAVVLYDSTQGVHAVRKALAPIFGLEPEQIRVIAPHVGGG
Mflv_0284|M.gilvum_PYR-GCK AQWTVRD-GTPAVTMYDSTQGVHVVRKTLAPIFGLEPEQMRVVAPHVGGG
MUL_4580|M.ulcerans_Agy99 AKWDGDR-----LTLWDKVQHINGARRSYARAFGMSEERVRLISPFVGGA
Mvan_2075|M.vanbaalenii_PYR-1 AQWDDGH-----LTVHDCTRWITGERRVLAAYLNIPEERIRVVSPLVGGA
MMAR_0080|M.marinum_M AQWDEDQ-----LTLWDAVQASATVPPVIATALGIDAHRIRIVAPHIGGG
Mb0380c|M.bovis_AF2122/97 ADLDPVTG---KLTLWTTSQAPHAHRTLYALVAGLPEHKIRVISPDIGGG
Rv0373c|M.tuberculosis_H37Rv ADLDPVTG---KLTLWTTSQAPHAHRTLYALVAGLPEHKIRVISPDIGGG
TH_0176|M.thermoresistible__bu ADLDPVTG---KLTLWSTSQAPHAHRTLYALVAGLPEHKIRVIAPDIGGG
MAB_2925c|M.abscessus_ATCC_199 GWQDERGR----YVLRTSSQVPFLVRDELCHVFGLAREQVRVFAPRVGGG
. .: : . .: .::*:.:* :**.
MSMEG_0684|M.smegmatis_MC2_155 FGSKGAPHAHNVLAILAAQRSNGRPVKLALTRQQMFSLVGYRTPTIQRIR
Mflv_0284|M.gilvum_PYR-GCK FGSKGAPHAHDVLTVMAAQRAGGRPVKLALTRQQMFSLVGYRTPTIQRVR
MUL_4580|M.ulcerans_Agy99 FGSAGRMWPHQLLTVFAARRMQ-LPVKLVLTRKQMYSSVGFRPTSRQRMA
Mvan_2075|M.vanbaalenii_PYR-1 FGSKSFLWMHVVLCAVAAREVG-RPVKLVLTRDQMFTSTGHRPRTEQQVA
MMAR_0080|M.marinum_M FGSKSFVWPHQTLAAAAARVTG-RPVKLHLRRADQFSSVGYQPWMEQTMR
Mb0380c|M.bovis_AF2122/97 FGNKVPIYPGYVCAIVASLLLD-KPVKWMEDRSENLTSTGFARDYIMVGE
Rv0373c|M.tuberculosis_H37Rv FGNKVPIYPGYVCAIVASLLLD-KPVKWMEDRSENLTSTGFARDYIMVGE
TH_0176|M.thermoresistible__bu FGNKVPIYPGYVCAIVGSLLLG-KPVKWVEDRSENLTSTGFARDYIMVGE
MAB_2925c|M.abscessus_ATCC_199 FGAKQEMLT-EDLVLLAVRVTG-APVVYEFTRSDQFTIAPCRHPMRVNVT
** . ** * : : .
MSMEG_0684|M.smegmatis_MC2_155 LGADHDGRITALVHDVVEQTSAVKEFAEQTAITSRKMYSAASRRTTHKLA
Mflv_0284|M.gilvum_PYR-GCK LGADRDGRLTALVHDAIGQTATVKEFAEQTAVTSRKMYAAANRRTSHRLA
MUL_4580|M.ulcerans_Agy99 IGADPSGRLMAMIHEGRTETAQYGIYEDGIVRSPQLTYQVPNMSSTYRVV
Mvan_2075|M.vanbaalenii_PYR-1 LVADAAGTIVSTEHHTLTETSTVAHFCEPAGVSSRILYQSPRLAVTHTVA
MMAR_0080|M.marinum_M LAADRTGRLIGIEHVAVNNTGMVDNYVEPATTATKGLYAADAIRTRQFVE
Mb0380c|M.bovis_AF2122/97 IAANRDGKILAIRSNVLADHGAFNAQAAPAKYP-AGFFGVFTGSYDIEAA
Rv0373c|M.tuberculosis_H37Rv IAANRDGKILAIRSNVLADHGAFNAQAAPAKYP-AGFFGVFTGSYDIEAA
TH_0176|M.thermoresistible__bu IAATRDGRILAIRSHVLADHGAFNGTAAPVKYP-AGFFGVFTGSYDLEAA
MAB_2925c|M.abscessus_ATCC_199 AAADGDGVLTALAVDVLSDAGAYGNHSVGVMFHGCNESMAQYRCANKRVD
* * : . : .
MSMEG_0684|M.smegmatis_MC2_155 ALDVAIPFWMRAPG---------ECPGTYAAEVAMDELAVACGLDPIELR
Mflv_0284|M.gilvum_PYR-GCK ALDVPVPFWMRAPG---------ECPGTYAAEVAMDELAVAAGIDPIQLR
MUL_4580|M.ulcerans_Agy99 PLDINLPWFMRGPG---------AVTGTFALESTMDDLAHRLGIDPIELR
Mvan_2075|M.vanbaalenii_PYR-1 RIHAPTPCFMRGPG---------EAPGLFALEMAMDELAYALGRDPLALR
MMAR_0080|M.marinum_M RVNLNVPTPMRAPL---------EGPGMWALESAMNELADEAGMDPLDLR
Mb0380c|M.bovis_AF2122/97 YCHMTAVYTNKAPGGVAYACSFRITEAVYFVERLVDCLAFELKMDPAELR
Rv0373c|M.tuberculosis_H37Rv YCHMTAVYTNKAPGGVAYACSFRITEAVYFVERLVDCLAFELKMDPAELR
TH_0176|M.thermoresistible__bu YCHMTAVYTNKAPGGVAYACSFRITEAVYLVERLVDCLAHELEMDPAELR
MAB_2925c|M.abscessus_ATCC_199 AQAVYTNNIPSGAFRG-----YGLGQVLFGIESALDELARNLGIDPFEFR
.. : * :: ** ** :*
MSMEG_0684|M.smegmatis_MC2_155 VRNDPEVD--------PETGKPWSGRHLVDCLRLGADRFGWAARD-PRPA
Mflv_0284|M.gilvum_PYR-GCK IRNDPPTD--------PETGKPWSGRHLVECLEVGAERFDWSSRD-PEPG
MUL_4580|M.ulcerans_Agy99 LRNEPRID--------QINGLPFSTRRLVDCIKQGSTDFGWHRRN-SIPG
Mvan_2075|M.vanbaalenii_PYR-1 MDNLADVD--------QTDGRPWSSAHLARCYTTGARRFGWDRRP-LQSR
MMAR_0080|M.marinum_M LANYAENS--------PADGRPWSSNRLREAYAEGARRYRWRERY-ERPR
Mb0380c|M.bovis_AF2122/97 LRNLLRPNQ---FPYQSKTGWVYDSGDYETTMRKAMNMIGYEALR-AEQK
Rv0373c|M.tuberculosis_H37Rv LRNLLRPNQ---FPYQSKTGWVYDSGDYETTMRKAMNMIGYEALR-AEQK
TH_0176|M.thermoresistible__bu LRNLLKPDQ---FPYTSKTGWVYDSGDYETTMRKAMAMVGYDELR-AEQA
MAB_2925c|M.abscessus_ATCC_199 RRNVIVPGDSFVSWEVEEDDLAFGSYGLDQCLDLAQDALARGAADPARNG
* . . :. .
MSMEG_0684|M.smegmatis_MC2_155 ARRDGEWLIGTGVAAATYPGKAMPG-----------NSARITYAAEGRYT
Mflv_0284|M.gilvum_PYR-GCK TRVLGDWLVGTGVASATYPGMAMPG-----------NAARITYSAEGHYD
MUL_4580|M.ulcerans_Agy99 ANRDGDSFVGIGMATAAYFTYRSK------------CAAIARISADGTAE
Mvan_2075|M.vanbaalenii_PYR-1 SLQRNGIRVGWGMATATYPGRRMP------------AACRVETDADGHVR
MMAR_0080|M.marinum_M --AEGPWAIGHGMATCTMGSFRFP------------SQARVRLRDDGTAV
Mb0380c|M.bovis_AF2122/97 QRRARGELMGIGMSFFTEAVGAGPRKDMDILGLGMADGCELRVHPTGKAV
Rv0373c|M.tuberculosis_H37Rv QRRARGELMGIGMSFFTEAVGAGPRKDMDILGLGMADGCELRVHPTGKAV
TH_0176|M.thermoresistible__bu ERRRRGELMGIGMSFFTEAVGAGPRKDMDILGLGMADGCELRVHPTGKAV
MAB_2925c|M.abscessus_ATCC_199 SDLPAGWKVGEGMAIAMIATVPPRG---------HVADARAELDARGRYR
:* *:: . *
MSMEG_0684|M.smegmatis_MC2_155 VQIGAADIGTGTWTTLSQIAADALACDLAAVDLQIGDTDLPEASV-AGGS
Mflv_0284|M.gilvum_PYR-GCK VHIGAADIGTGTWTTLSQIAADALDCDLAAITLQIGDTDLPAASV-AGGS
MUL_4580|M.ulcerans_Agy99 VCSAASDMGPGTYTSMTQIAADALGLPLQRVRFSLGDSRFPPAPA-HSAS
Mvan_2075|M.vanbaalenii_PYR-1 FSAATHEIGTGVRTVMTQLAADTTGLPLSQVSFESGDSSFPDAPY-SGAS
MMAR_0080|M.marinum_M VEANVHDIGTGVQTILCQIAAAELGISSDRVEILWGDTDLPTAGP-AYGS
Mb0380c|M.bovis_AF2122/97 LRLSVQTQGQGHETTFAQIVAEELGIAPDDIEVVHGDTDQTPFGLGTYGS
Rv0373c|M.tuberculosis_H37Rv LRLSVQTQGQGHETTFAQIVAEELGIAPDDIEVVHGDTDQTPFGLGTYGS
TH_0176|M.thermoresistible__bu VRLSVQTQGQGHETTFAQIVAEELGIPPDDIEVVHGDTDQTPFGLGTYGS
MAB_2925c|M.abscessus_ATCC_199 ISVGSAEFGNGTTTVHAQLAAGVLGCAPDQVYIHQADTDAVSHDTGAFGS
. * * * *:.* : . .*: .*
MSMEG_0684|M.smegmatis_MC2_155 SGITSWGSAIVAAARQFRA-------------------DHGDPPAVG---
Mflv_0284|M.gilvum_PYR-GCK SGMTSWGTTIVLAAQQFRR-------------------DHGNPPVIG---
MUL_4580|M.ulcerans_Agy99 RTTASVGSAVFTVANMLHDKFIRAAV--TDPQSPLAGLRPEDVMVTNGHM
Mvan_2075|M.vanbaalenii_PYR-1 QTSASVGSAVFAAAQEWRG-------------------RADALPAT----
MMAR_0080|M.marinum_M STTMSVGSAVAAAARHVVN--------------QLAALGIDDPAATG---
Mb0380c|M.bovis_AF2122/97 RSTPVSGGAAALVARKVRDKAKIIASGMLEVSVADLQWEKGKFHVKGDPS
Rv0373c|M.tuberculosis_H37Rv RSTPVSGGAAALVARKVRDKAKIIASGMLEVSVADLQWEKGKFHVKGDPS
TH_0176|M.thermoresistible__bu RSTPVSGAAAALVARKVRDKAKIIASGMLEASIADLEWEKGSFRVKGDPS
MAB_2925c|M.abscessus_ATCC_199 AGTTVAGLAVYRAAELLRDQIIRFAATKLAVPQSRCVLEPDGVRAGADLL
* : .*. .
MSMEG_0684|M.smegmatis_MC2_155 --------------------------ATTVAQSPENPDADGVTMQSFGAH
Mflv_0284|M.gilvum_PYR-GCK --------------------------ATTVTEAPD-PENDEVTVQSFGAH
MUL_4580|M.ulcerans_Agy99 FSTVATDGPARGESYQDLLRRRGWPALESEHTWTPDDAGKRYSTYAYGAV
Mvan_2075|M.vanbaalenii_PYR-1 -----ADGPGR-------------AAALSFTTASGPGPDVDTVSRSFGAH
MMAR_0080|M.marinum_M --IAEVVGEGR--------------FSTPGGGTNLDGEGTPFAIRAWGAV
Mb0380c|M.bovis_AF2122/97 AAVTIADIAMRAHG------AGDLPEGIEGGLDAEVCYNPSNLTYPYGAY
Rv0373c|M.tuberculosis_H37Rv AAVTIADIAMRAHG------AGDLPEGIEGGLDAEVCYNPSNLTYPYGAY
TH_0176|M.thermoresistible__bu ASVTIQDIAMRAHG------AGDLPDGIEGGLDAQICYNPSNLTYPYGAY
MAB_2925c|M.abscessus_ATCC_199 G-------------------LTEIAASAPEPLSAVGHHEGTPRSVAFNVQ
.:..
MSMEG_0684|M.smegmatis_MC2_155 FVEAHVNADTGEIRVPRMLGVFSVGRVINARTLRSQLIGGMTMGLSMALH
Mflv_0284|M.gilvum_PYR-GCK FVEAHVNRHTGEIRVPRMLGVFSIGRAINPRTLRSQLIGGMTMGLSMALH
MUL_4580|M.ulcerans_Agy99 FAEVAIDALLPTVRIRRIYACYDAGRVINPKLAHSQAIGGMIGGIGMALL
Mvan_2075|M.vanbaalenii_PYR-1 FCEVEVDEQIGRATVTRWVAVMDCGRVLNPKLATNQVMGGILFGLGMALM
MMAR_0080|M.marinum_M FVEVGVDRDLGLIRLRHAVGVYSAGRILNPVTARAQMIGGIIWGWGKATL
Mb0380c|M.bovis_AF2122/97 FCVVDIDPGTAVVKVRRFLAVDDCGTRINPMIIEGQVHGGIVDGIGMALM
Rv0373c|M.tuberculosis_H37Rv FCVVDIDPGTAVVKVRRFLAVDDCGTRINPMIIEGQVHGGIVDGIGMALM
TH_0176|M.thermoresistible__bu FCVVDVDPGTAVVKVRRFLAVDDCGTRINPMIIEGQVHGGIVDGIGMALM
MAB_2925c|M.abscessus_ATCC_199 AFRVAVRPQTGTVRILQSVQTADAGVVVNPQQCRGQVEGGVAQAIGSALY
. : : : . * :*. * **: . . *
MSMEG_0684|M.smegmatis_MC2_155 EESVRDPRFGHVVTQDLATYHISAHADVA-DIDAIWLDEADEYLNPMGSR
Mflv_0284|M.gilvum_PYR-GCK EESVRDPRFGHVVTQDFATYHISSHADVV-DIDAIWLDEVDEHLNPMGSR
MUL_4580|M.ulcerans_Agy99 EGTELDYRDGRVVNANISDYLVPINADVP-VLDASFLPAQDTIADPIGVK
Mvan_2075|M.vanbaalenii_PYR-1 EQVPHDPHTGRLLG----EYYVPTHADRP-AFDISFVDSSDYGLDPIGVR
MMAR_0080|M.marinum_M EASTQEPTYGRWLAKNLSNVALPVNADIPAAIDVSFIDEFDSAASLIGAR
Mb0380c|M.bovis_AF2122/97 EMIAFD-EDGNCLGGSLMDYLIPTALEVP-HLETGHTVTP-SPHHPIGAK
Rv0373c|M.tuberculosis_H37Rv EMIAFD-EDGNCLGGSLMDYLIPTALEVP-HLETGHTVTP-SPHHPIGAK
TH_0176|M.thermoresistible__bu QMIGFD-EDGNCLGGSLMDYLIPTAMEVP-TLETGFTVTP-SPHHPIGAK
MAB_2925c|M.abscessus_ATCC_199 EEILVH--DGFVSTTALRNYHIPRLVDVP--DTEVYFADTYDTFGPLGAK
: . * :. : :* :
MSMEG_0684|M.smegmatis_MC2_155 GAGEIGIVGSASAVVNAVYHATG---VRVRNLPVTVDKVLAGLPKLV---
Mflv_0284|M.gilvum_PYR-GCK GAGEIGIVGSAAAVVNAVYHATG---IRVRDLPVTLDKVLAGLP------
MUL_4580|M.ulcerans_Agy99 GLGELVIIAVPAAIANAVFNATG---KRVTDLPIRVEQLL----------
Mvan_2075|M.vanbaalenii_PYR-1 GIGEIGVCGVAGAIASAIHHATG---KWLRQLPITLEALLGPRDDDRED-
MMAR_0080|M.marinum_M GIGELGATGVDAAVAAAVYDAVG---VRVRELPILPWQVLS---------
Mb0380c|M.bovis_AF2122/97 GIGESATVGSPPAVVNAVVDALAPFGVRHADMPLTPSRVWEAMQGRATPP
Rv0373c|M.tuberculosis_H37Rv GIGESATVGSPPAVVNAVVDALAPFGVRHADMPLTPSRVWEAMQGRATPP
TH_0176|M.thermoresistible__bu GIGESATVGSPPAVVNAVVDALAPFGVRHVDMPLTPSRVWEAMQGRAQPS
MAB_2925c|M.abscessus_ATCC_199 SMSESPYNPVAPALANAIRDAVG---VRVYRLPMNAARVWRVINDSGGGE
. .* *:. *: .* . :*: :
MSMEG_0684|M.smegmatis_MC2_155 --
Mflv_0284|M.gilvum_PYR-GCK --
MUL_4580|M.ulcerans_Agy99 --
Mvan_2075|M.vanbaalenii_PYR-1 --
MMAR_0080|M.marinum_M --
Mb0380c|M.bovis_AF2122/97 I-
Rv0373c|M.tuberculosis_H37Rv I-
TH_0176|M.thermoresistible__bu I-
MAB_2925c|M.abscessus_ATCC_199 SS