For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDFAYVIAGSEATGGAGLQADLRTFQQLGAYGVGTVTCIVSFDPKAGWSHRFVPVEAQVVADQIEAATSA YDLHTVKIGMLGTPATIDVVAEGLRGQQWRHIVVDPVLICKGQEPGAALDTDTALRREILPLATVTTPNL FEARTLSGMDEITTVDDLIEAARRIADLGPRHVVVKGGVEFPGSDAVDVLFDRDSDGRNAQVLRAAKVGE ARVAGAGCTLAAAITAELAKGADVPEAVLRAKEFTTAGIVARVGGNAPFDAVWQGGQ*
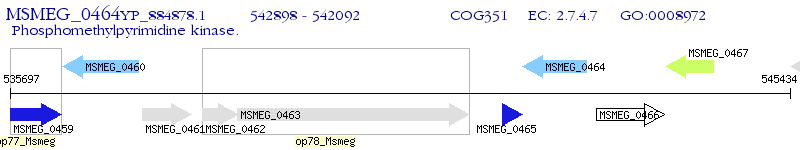
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0464 | - | - | 100% (268) | phosphomethylpyrimidine kinase |
| M. smegmatis MC2 155 | MSMEG_0825 | thiD | 1e-28 | 32.82% (262) | phosphomethylpyrimidine kinase |
| M. smegmatis MC2 155 | MSMEG_3947 | - | 8e-06 | 30.77% (130) | 6-phosphofructokinase isozyme 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0430c | thiD | 2e-26 | 36.10% (241) | phosphomethylpyrimidine kinase |
| M. gilvum PYR-GCK | Mflv_0181 | - | 1e-30 | 36.73% (245) | phosphomethylpyrimidine kinase |
| M. tuberculosis H37Rv | Rv0422c | thiD | 2e-26 | 36.10% (241) | phosphomethylpyrimidine kinase |
| M. leprae Br4923 | MLBr_00295 | thiD | 5e-30 | 36.51% (241) | phosphomethylpyrimidine kinase |
| M. abscessus ATCC 19977 | MAB_4197 | - | 2e-28 | 34.44% (270) | phosphomethylpyrimidine kinase |
| M. marinum M | MMAR_0734 | thiD | 1e-29 | 37.80% (254) | phosphomethylpyrimidine kinase ThiD |
| M. avium 104 | MAV_4732 | thiD | 6e-30 | 35.96% (267) | phosphomethylpyrimidine kinase |
| M. thermoresistible (build 8) | TH_1214 | thiD | 3e-35 | 42.39% (243) | PROBABLE PHOSPHOMETHYLPYRIMIDINE KINASE THID (HMP-PHOSPHATE |
| M. ulcerans Agy99 | MUL_3599 | thiD | 7e-30 | 37.80% (254) | phosphomethylpyrimidine kinase |
| M. vanbaalenii PYR-1 | Mvan_0724 | - | 2e-31 | 36.84% (266) | phosphomethylpyrimidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0734|M.marinum_M ---------MSWLPLAPPGATPLRALTIAGSDSGGGAGIQADMRTMALLG
MUL_3599|M.ulcerans_Agy99 ---------MSWLPLAPPGATPLRALTIAGSDSGGGAGIQADMRTMALLG
Mb0430c|M.bovis_AF2122/97 -------------------MTPPRVLSIAGSDSGGGAGIQADMRTMALLG
Rv0422c|M.tuberculosis_H37Rv -------------------MTPPRVLSIAGSDSGGGAGIQADMRTMALLG
MAV_4732|M.avium_104 -----MTGPPPTLPLAPPGTTPRRVLSIAGSDSGGGAGIQADLRTMAMLG
MLBr_00295|M.leprae_Br4923 ---------MNYLPAPPPGTTPLRVLSIAGSDSGGGAGIQADMRTMTVLG
Mflv_0181|M.gilvum_PYR-GCK MTHSTTATTSSFLPLTPAGQTPLRVLTIAGSDSGGGAGIQADMRTFALLG
Mvan_0724|M.vanbaalenii_PYR-1 ---------MTFLPLTPPGQTPLRVMTIAGSDSGGGAGIQADMRTFALLG
TH_1214|M.thermoresistible__bu ---------VTYLPLPEPGQTPLRVLTIAGSDSGGGAGIQADLRTFAMLG
MAB_4197|M.abscessus_ATCC_1997 -------MRSPFLPLPEPGTTPVRAMTIAGSDSGGGAGIQADMRTMALLG
MSMEG_0464|M.smegmatis_MC2_155 --------------------MVDFAYVIAGSEATGGAGLQADLRTFQQLG
. ****:: ****:***:**: **
MMAR_0734|M.marinum_M VHACVAVTAVTVQNTLG--VKGFHETPEDVVAGQITAVVEDIGIQAAKTG
MUL_3599|M.ulcerans_Agy99 VHACVVVTAVTVQNTLG--VKGFHETPEDVVAGQITAVVEDIGIQAAKTG
Mb0430c|M.bovis_AF2122/97 VHACVAVTAVTVQNTLG--VKDIHEVPNDVVAGQIEAVVTDIGVQAAKTG
Rv0422c|M.tuberculosis_H37Rv VHACVAVTAVTVQNTLG--VKDIHEVPNDVVAGQIEAVVTDIGVQAAKTG
MAV_4732|M.avium_104 VHACVAVTAVTAQNTLG--VNSFHEVPADVVAGQIEAVAGDIGIQAAKTG
MLBr_00295|M.leprae_Br4923 VHACTAVTAVTIQNTLG--VEGFHEIPAEIVASQIKAVVTDIGIQSAKTG
Mflv_0181|M.gilvum_PYR-GCK VHSLVAVTAVTVQNSLG--VKGFHEIPLDVIAGQIEAVASDIGLQAAKTG
Mvan_0724|M.vanbaalenii_PYR-1 LHGLVAVTAVTVQNSVG--VKGFHEIPLDVVAGQIEAVASDIGLQAAKTG
TH_1214|M.thermoresistible__bu VHGLVAVAAVTVQNSVG--VSGFHEIPVQVVADQIVAVASDIGLQAAKTG
MAB_4197|M.abscessus_ATCC_1997 VHGCVAVTAVTVQNSLG--VKDFHEIPPPIVAAQMEVVITDIGIQAAKTG
MSMEG_0464|M.smegmatis_MC2_155 AYGVGTVTCIVSFDPKAGWSHRFVPVEAQVVADQIEAATSAYDLHTVKIG
:. .*:.:. :. . : ::* *: .. .:::.* *
MMAR_0734|M.marinum_M MLASPRIIDAVAATWRQ--LEVAVPLVVDPVCASMHGDALLAASGLESLR
MUL_3599|M.ulcerans_Agy99 MLASPRIIDAVAATWRQ--LEVTVPLVVDPVCASMHGDALLAASGLESLR
Mb0430c|M.bovis_AF2122/97 MLASSRIVATVAATWRR--LELSVPLVVDPVCASMHGDPLLAPSALDSLR
Rv0422c|M.tuberculosis_H37Rv MLASSRIVATVAATWRR--LELSVPLVVDPVCASMHGDPLLAPSALDSLR
MAV_4732|M.avium_104 MLASSPIIAEVAATWRR--LGLSVPLVVDPVCASMHGDPLLAADALDTLR
MLBr_00295|M.leprae_Br4923 MLASSSIIAAVAETWLR--LELTTPLVVDPVCASMHGDPLLTGEAMDSLR
Mflv_0181|M.gilvum_PYR-GCK MLASSEIIETIADTWVAQGLSGTVPLIVDPVCASMHGDPLLHPSALNALK
Mvan_0724|M.vanbaalenii_PYR-1 MLASSEIIETIADTWVAQGLHGSVPLVVDPVCASMHGDPLLHPSALNALR
TH_1214|M.thermoresistible__bu MLASSEIIETVAETARGQGIGTAVPLVVDPVCASMHGDPLLHMSALDALR
MAB_4197|M.abscessus_ATCC_1997 MLASAPIIEAISESWSN--LAPSVPLVVDPVCASMHGDPLLHPSALDALR
MSMEG_0464|M.smegmatis_MC2_155 MLGTPATIDVVAEGLRG---QQWRHIVVDPVLICKGQEPGAALDTDTALR
**.:. : :: ::**** . :. . :*:
MMAR_0734|M.marinum_M EQLFPLATLVTPNLDEVRLLVGIDVID-TQSQHAAARALHGLGPRWVLVK
MUL_3599|M.ulcerans_Agy99 EQLFPLATLVTPNLDEVRLLVGIDVID-TQSQHAAARALHGLGPRWVLVK
Mb0430c|M.bovis_AF2122/97 GQLFPLATLLTPNLDEARLLVDIEVVD-AESQRAAAKALHALGPQWVLVK
Rv0422c|M.tuberculosis_H37Rv GQLFPLATLLTPNLDEARLLVDIEVVD-AESQRAAAKALHALGPQWVLVK
MAV_4732|M.avium_104 GQLFPLATLVTPNLDEVRLLVDIDVTD-AASQRDAAKALHALGPQWVLVK
MLBr_00295|M.leprae_Br4923 DRLFPLATVVTPNLDEVRLLVGIDVVD-TESQRAAAMALHALGPQWALVK
Mflv_0181|M.gilvum_PYR-GCK EKLFPLATLVTPNLDEVGLLTGIEVVD-EDSQRDAARALHALGPSWALVK
Mvan_0724|M.vanbaalenii_PYR-1 DKLFPLATLVTPNLDEVRLLVDIDVVD-EESQRAAARALHALGPQWALVK
TH_1214|M.thermoresistible__bu TKLFPIATLVTPNLDEVRLLVGVDVVD-EASQREAAKALHALGPKWVLVK
MAB_4197|M.abscessus_ATCC_1997 TRLFPLATLVTPNLDEVRLLVDIDVVD-EDSQKEAARKLHALGPQWALVK
MSMEG_0464|M.smegmatis_MC2_155 REILPLATVTTPNLFEARTLSGMDEITTVDDLIEAARRIADLGPRHVVVK
.::*:**: **** *. * .:: . ** : *** .:**
MMAR_0734|M.marinum_M GGHLRSSTRSCDLLYDG----TDFFEFDAPRVATGNDHGGGDTLAAATAC
MUL_3599|M.ulcerans_Agy99 GGHLRSSTRSCDLLYDG----TDFFEFDAPRVATGNDHGGGDTLAAATAC
Mb0430c|M.bovis_AF2122/97 GGHLRSSDGSCDLLYDG----VSCYQFDAQRLPTGDDHGGGDTLATAIAA
Rv0422c|M.tuberculosis_H37Rv GGHLRSSDGSCDLLYDG----VSCYQFDAQRLPTGDDHGGGDTLATAIAA
MAV_4732|M.avium_104 GGHLRSSQRSCDLLYDG----DSCYEFDAHRLNTGNDHGGGDTLASAVAC
MLBr_00295|M.leprae_Br4923 GGHLRSSDRSCDLLYGGSSAGVAFHEFDAPRVQTGNDHGGGDTLAAAVSC
Mflv_0181|M.gilvum_PYR-GCK GGHLRSSAQSPDLLFDG----TDFHRFDSTRIDTTHDHGAGDTLAAATAS
Mvan_0724|M.vanbaalenii_PYR-1 GGHLRSSSHSPDLLFDG----TDFHAFDSTRIDTVHDHGAGDTLAAAIAC
TH_1214|M.thermoresistible__bu GGHLRSAPHSPDLLFDG----AEFHRFDAPRVDTGHDHGAGDTLAAATAA
MAB_4197|M.abscessus_ATCC_1997 GGHLRSSDTSTDLLFDG----TEFHYFPVERVDTGHDHGAGDTLAASISC
MSMEG_0464|M.smegmatis_MC2_155 GGVEFPGSDAVDVLFDRDSDGRNAQVLRAAKVGEARVAGAGCTLAAAITA
** .. : *:*:. : :: *.* ***:: :.
MMAR_0734|M.marinum_M ALAHGFTVPDAVSFAKKWVTECIRAAYPLGRGHGPVSPLFRLS---
MUL_3599|M.ulcerans_Agy99 ALAHGFTVPDAVSFAKKWVTECIRAAYPLGRGHGPVSPLFRLS---
Mb0430c|M.bovis_AF2122/97 ALAHGFTVPDAVDFGKRWVTECLRAAYPLGRGHGPVSPLFRLS---
Rv0422c|M.tuberculosis_H37Rv ALAHGFTVPDAVDFGKRWVTECLRAAYPLGRGHGPVSPLFRLS---
MAV_4732|M.avium_104 ALAHGFTVPDAVAFGKRWVTECLRASYPLGHGHGPVSALFRLSGDG
MLBr_00295|M.leprae_Br4923 ALAHGYTVPDAVGFGKRWVTECLRDAYPLGRGHGPVSPLFQRT---
Mflv_0181|M.gilvum_PYR-GCK ALAHGYSVPDAVAFAKRWVTECIRSAYPLGQGHGPVSALFRLAP--
Mvan_0724|M.vanbaalenii_PYR-1 ALAHGYPVPDAVAFGKHWVTECLRAAYPLGHGHGPVSALFRLTT--
TH_1214|M.thermoresistible__bu ALAHGHSVPEAVEFAKRWITECLRAAYPLGRGHGPVNPMFRLRG--
MAB_4197|M.abscessus_ATCC_1997 ALAHGLPVTEAVAFGKQWITECLKAAYPLGHGHGPVSALFRLDA--
MSMEG_0464|M.smegmatis_MC2_155 ELAKGADVPEAVLRAKEFTTAGIVARVG---GNAPFDAVWQGGQ--
**:* *.:** .*.: * : *:.*...:::