For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRSPFLPLPEPGTTPVRAMTIAGSDSGGGAGIQADMRTMALLGVHGCVAVTAVTVQNSLGVKDFHEIPPP IVAAQMEVVITDIGIQAAKTGMLASAPIIEAISESWSNLAPSVPLVVDPVCASMHGDPLLHPSALDALRT RLFPLATLVTPNLDEVRLLVDIDVVDEDSQKEAARKLHALGPQWALVKGGHLRSSDTSTDLLFDGTEFHY FPVERVDTGHDHGAGDTLAASISCALAHGLPVTEAVAFGKQWITECLKAAYPLGHGHGPVSALFRLDA
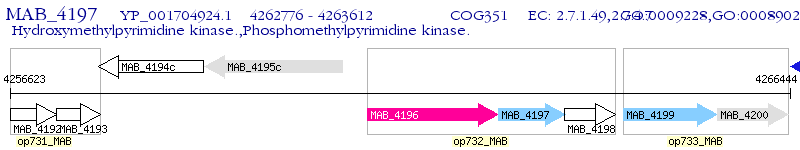
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4197 | - | - | 100% (278) | phosphomethylpyrimidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0430c | thiD | 1e-111 | 72.62% (263) | phosphomethylpyrimidine kinase |
| M. gilvum PYR-GCK | Mflv_0181 | - | 1e-115 | 73.55% (276) | phosphomethylpyrimidine kinase |
| M. tuberculosis H37Rv | Rv0422c | thiD | 1e-111 | 72.62% (263) | phosphomethylpyrimidine kinase |
| M. leprae Br4923 | MLBr_00295 | thiD | 1e-113 | 70.91% (275) | phosphomethylpyrimidine kinase |
| M. marinum M | MMAR_0734 | thiD | 1e-111 | 71.69% (272) | phosphomethylpyrimidine kinase ThiD |
| M. avium 104 | MAV_4732 | thiD | 1e-115 | 72.16% (273) | phosphomethylpyrimidine kinase |
| M. smegmatis MC2 155 | MSMEG_0825 | thiD | 1e-117 | 73.06% (271) | phosphomethylpyrimidine kinase |
| M. thermoresistible (build 8) | TH_1214 | thiD | 1e-114 | 73.36% (274) | PROBABLE PHOSPHOMETHYLPYRIMIDINE KINASE THID (HMP-PHOSPHATE |
| M. ulcerans Agy99 | MUL_3599 | thiD | 1e-111 | 71.32% (272) | phosphomethylpyrimidine kinase |
| M. vanbaalenii PYR-1 | Mvan_0724 | - | 1e-124 | 79.56% (274) | phosphomethylpyrimidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0825|M.smegmatis_MC2_155 ---------MKLLPLTPPGQTPTRVMTIAGTDSGGGAGIQADLRTFAMLG
TH_1214|M.thermoresistible__bu ---------VTYLPLPEPGQTPLRVLTIAGSDSGGGAGIQADLRTFAMLG
Mflv_0181|M.gilvum_PYR-GCK MTHSTTATTSSFLPLTPAGQTPLRVLTIAGSDSGGGAGIQADMRTFALLG
Mvan_0724|M.vanbaalenii_PYR-1 ---------MTFLPLTPPGQTPLRVMTIAGSDSGGGAGIQADMRTFALLG
MAB_4197|M.abscessus_ATCC_1997 -------MRSPFLPLPEPGTTPVRAMTIAGSDSGGGAGIQADMRTMALLG
Mb0430c|M.bovis_AF2122/97 -------------------MTPPRVLSIAGSDSGGGAGIQADMRTMALLG
Rv0422c|M.tuberculosis_H37Rv -------------------MTPPRVLSIAGSDSGGGAGIQADMRTMALLG
MAV_4732|M.avium_104 -----MTGPPPTLPLAPPGTTPRRVLSIAGSDSGGGAGIQADLRTMAMLG
MLBr_00295|M.leprae_Br4923 ---------MNYLPAPPPGTTPLRVLSIAGSDSGGGAGIQADMRTMTVLG
MMAR_0734|M.marinum_M ---------MSWLPLAPPGATPLRALTIAGSDSGGGAGIQADMRTMALLG
MUL_3599|M.ulcerans_Agy99 ---------MSWLPLAPPGATPLRALTIAGSDSGGGAGIQADMRTMALLG
** *.::***:***********:**:::**
MSMEG_0825|M.smegmatis_MC2_155 VHGCVAVAAVTVQNSVGVKGFHEVPVDIIAGQIQVVAEDIGIQAAKTGML
TH_1214|M.thermoresistible__bu VHGLVAVAAVTVQNSVGVSGFHEIPVQVVADQIVAVASDIGLQAAKTGML
Mflv_0181|M.gilvum_PYR-GCK VHSLVAVTAVTVQNSLGVKGFHEIPLDVIAGQIEAVASDIGLQAAKTGML
Mvan_0724|M.vanbaalenii_PYR-1 LHGLVAVTAVTVQNSVGVKGFHEIPLDVVAGQIEAVASDIGLQAAKTGML
MAB_4197|M.abscessus_ATCC_1997 VHGCVAVTAVTVQNSLGVKDFHEIPPPIVAAQMEVVITDIGIQAAKTGML
Mb0430c|M.bovis_AF2122/97 VHACVAVTAVTVQNTLGVKDIHEVPNDVVAGQIEAVVTDIGVQAAKTGML
Rv0422c|M.tuberculosis_H37Rv VHACVAVTAVTVQNTLGVKDIHEVPNDVVAGQIEAVVTDIGVQAAKTGML
MAV_4732|M.avium_104 VHACVAVTAVTAQNTLGVNSFHEVPADVVAGQIEAVAGDIGIQAAKTGML
MLBr_00295|M.leprae_Br4923 VHACTAVTAVTIQNTLGVEGFHEIPAEIVASQIKAVVTDIGIQSAKTGML
MMAR_0734|M.marinum_M VHACVAVTAVTVQNTLGVKGFHETPEDVVAGQITAVVEDIGIQAAKTGML
MUL_3599|M.ulcerans_Agy99 VHACVVVTAVTVQNTLGVKGFHETPEDVVAGQITAVVEDIGIQAAKTGML
:*. ..*:*** **::**..:** * ::* *: .* ***:*:******
MSMEG_0825|M.smegmatis_MC2_155 ASAEIIGAVAETWRG--LGTEAPLVVDPVCASMHGDPLLHPSALDSVRNE
TH_1214|M.thermoresistible__bu ASSEIIETVAETARGQGIGTAVPLVVDPVCASMHGDPLLHMSALDALRTK
Mflv_0181|M.gilvum_PYR-GCK ASSEIIETIADTWVAQGLSGTVPLIVDPVCASMHGDPLLHPSALNALKEK
Mvan_0724|M.vanbaalenii_PYR-1 ASSEIIETIADTWVAQGLHGSVPLVVDPVCASMHGDPLLHPSALNALRDK
MAB_4197|M.abscessus_ATCC_1997 ASAPIIEAISESWSN--LAPSVPLVVDPVCASMHGDPLLHPSALDALRTR
Mb0430c|M.bovis_AF2122/97 ASSRIVATVAATWRR--LELSVPLVVDPVCASMHGDPLLAPSALDSLRGQ
Rv0422c|M.tuberculosis_H37Rv ASSRIVATVAATWRR--LELSVPLVVDPVCASMHGDPLLAPSALDSLRGQ
MAV_4732|M.avium_104 ASSPIIAEVAATWRR--LGLSVPLVVDPVCASMHGDPLLAADALDTLRGQ
MLBr_00295|M.leprae_Br4923 ASSSIIAAVAETWLR--LELTTPLVVDPVCASMHGDPLLTGEAMDSLRDR
MMAR_0734|M.marinum_M ASPRIIDAVAATWRQ--LEVAVPLVVDPVCASMHGDALLAASGLESLREQ
MUL_3599|M.ulcerans_Agy99 ASPRIIDAVAATWRQ--LEVTVPLVVDPVCASMHGDALLAASGLESLREQ
**. *: :: : : .**:***********.** ..::::: .
MSMEG_0825|M.smegmatis_MC2_155 LFPIASLVTPNLDEVRLITGIEVVDEATQRDAAKALHALGPKWALVKGGH
TH_1214|M.thermoresistible__bu LFPIATLVTPNLDEVRLLVGVDVVDEASQREAAKALHALGPKWVLVKGGH
Mflv_0181|M.gilvum_PYR-GCK LFPLATLVTPNLDEVGLLTGIEVVDEDSQRDAARALHALGPSWALVKGGH
Mvan_0724|M.vanbaalenii_PYR-1 LFPLATLVTPNLDEVRLLVDIDVVDEESQRAAARALHALGPQWALVKGGH
MAB_4197|M.abscessus_ATCC_1997 LFPLATLVTPNLDEVRLLVDIDVVDEDSQKEAARKLHALGPQWALVKGGH
Mb0430c|M.bovis_AF2122/97 LFPLATLLTPNLDEARLLVDIEVVDAESQRAAAKALHALGPQWVLVKGGH
Rv0422c|M.tuberculosis_H37Rv LFPLATLLTPNLDEARLLVDIEVVDAESQRAAAKALHALGPQWVLVKGGH
MAV_4732|M.avium_104 LFPLATLVTPNLDEVRLLVDIDVTDAASQRDAAKALHALGPQWVLVKGGH
MLBr_00295|M.leprae_Br4923 LFPLATVVTPNLDEVRLLVGIDVVDTESQRAAAMALHALGPQWALVKGGH
MMAR_0734|M.marinum_M LFPLATLVTPNLDEVRLLVGIDVIDTQSQHAAARALHGLGPRWVLVKGGH
MUL_3599|M.ulcerans_Agy99 LFPLATLVTPNLDEVRLLVGIDVIDTQSQHAAARALHGLGPRWVLVKGGH
***:*:::******. *:..::* * :*: ** **.*** *.******
MSMEG_0825|M.smegmatis_MC2_155 LRTSAQSPDLLYNGTDF----YEFGAERIDTGHDHGAGDTLAAAAACALA
TH_1214|M.thermoresistible__bu LRSAPHSPDLLFDGAEF----HRFDAPRVDTGHDHGAGDTLAAATAAALA
Mflv_0181|M.gilvum_PYR-GCK LRSSAQSPDLLFDGTDF----HRFDSTRIDTTHDHGAGDTLAAATASALA
Mvan_0724|M.vanbaalenii_PYR-1 LRSSSHSPDLLFDGTDF----HAFDSTRIDTVHDHGAGDTLAAAIACALA
MAB_4197|M.abscessus_ATCC_1997 LRSSDTSTDLLFDGTEF----HYFPVERVDTGHDHGAGDTLAASISCALA
Mb0430c|M.bovis_AF2122/97 LRSSDGSCDLLYDGVSC----YQFDAQRLPTGDDHGGGDTLATAIAAALA
Rv0422c|M.tuberculosis_H37Rv LRSSDGSCDLLYDGVSC----YQFDAQRLPTGDDHGGGDTLATAIAAALA
MAV_4732|M.avium_104 LRSSQRSCDLLYDGDSC----YEFDAHRLNTGNDHGGGDTLASAVACALA
MLBr_00295|M.leprae_Br4923 LRSSDRSCDLLYGGSSAGVAFHEFDAPRVQTGNDHGGGDTLAAAVSCALA
MMAR_0734|M.marinum_M LRSSTRSCDLLYDGTDF----FEFDAPRVATGNDHGGGDTLAAATACALA
MUL_3599|M.ulcerans_Agy99 LRSSTRSCDLLYDGTDF----FEFDAPRVATGNDHGGGDTLAAATACALA
**:: * ***:.* . . * *: * .***.*****:: :.***
MSMEG_0825|M.smegmatis_MC2_155 HGMDMPDAVAFAKGWVTECLRAAYPLGHGHGPVSALFRLHHGS
TH_1214|M.thermoresistible__bu HGHSVPEAVEFAKRWITECLRAAYPLGRGHGPVNPMFRLRG--
Mflv_0181|M.gilvum_PYR-GCK HGYSVPDAVAFAKRWVTECIRSAYPLGQGHGPVSALFRLAP--
Mvan_0724|M.vanbaalenii_PYR-1 HGYPVPDAVAFGKHWVTECLRAAYPLGHGHGPVSALFRLTT--
MAB_4197|M.abscessus_ATCC_1997 HGLPVTEAVAFGKQWITECLKAAYPLGHGHGPVSALFRLDA--
Mb0430c|M.bovis_AF2122/97 HGFTVPDAVDFGKRWVTECLRAAYPLGRGHGPVSPLFRLS---
Rv0422c|M.tuberculosis_H37Rv HGFTVPDAVDFGKRWVTECLRAAYPLGRGHGPVSPLFRLS---
MAV_4732|M.avium_104 HGFTVPDAVAFGKRWVTECLRASYPLGHGHGPVSALFRLSGDG
MLBr_00295|M.leprae_Br4923 HGYTVPDAVGFGKRWVTECLRDAYPLGRGHGPVSPLFQRT---
MMAR_0734|M.marinum_M HGFTVPDAVSFAKKWVTECIRAAYPLGRGHGPVSPLFRLS---
MUL_3599|M.ulcerans_Agy99 HGFTVPDAVSFAKKWVTECIRAAYPLGRGHGPVSPLFRLS---
** :.:** *.* *:***:: :****:*****..:*: