For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPIRTLAVAGRLPDDGGVTALSTALPVVDLGDDAAVVTDQLRRVAHEVGFFYLVGHGVPQDLVDRVLTAA RRLFALPQADKDAIAMVNSPHFRGYTRLGGELTGGQVDWREQIDIGPEREPLDDPAEPYLRLQGPNQWPA ALPELPAVIAEWDAALAQVGRALLRHWAVALGNPADVFDEAFADTPATLIKIVRYPAQAETSQGVGAHRD AGVLTLLLAEPGSRGLQVRGPDGVFIDVDPLPGAFIVNIGEMLEIASGGYLRATEHRVQVSASERISVPY FFNPRLDAAIPVLALPDELAAQARGVTADPSNDRIYSVYGRNAWKSRVRAHPDVAAAHGYV
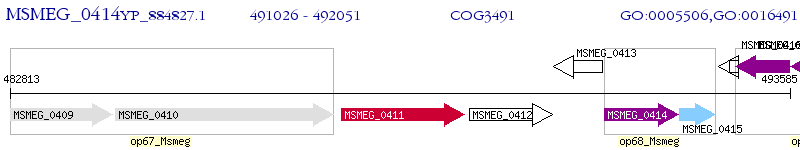
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0414 | - | - | 100% (341) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_4008 | - | 5e-30 | 30.23% (301) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_4685 | - | 1e-27 | 32.04% (309) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2580 | - | 5e-23 | 32.86% (283) | 2OG-Fe(II) oxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4434c | - | 1e-117 | 63.56% (343) | putative iron/ascorbate dependent oxidoreductase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4913 | - | 1e-128 | 72.39% (326) | oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| M. thermoresistible (build 8) | TH_4524 | - | 9e-43 | 40.70% (285) | PUTATIVE 2OG-Fe(II) oxygenase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4054 | - | 1e-24 | 33.33% (303) | 2OG-Fe(II) oxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0414|M.smegmatis_MC2_155 MPIRTLAVAGRLPDDGGVTALSTALPVVDLGDDAAVVTDQLRRVAHEVGF
MAV_4913|M.avium_104 MSNDMCPVPG-----------VTAVPGVDLRADPDVLRDGLREAAHRVGF
MAB_4434c|M.abscessus_ATCC_199 MAVKTVAPKRLASNHG----RVAELPVIDLSSDSNQLRAALREVAHEVGF
Mflv_2580|M.gilvum_PYR-GCK MSVIATVDLS---------------RWREGGPAAEDVAAQVDAGLQRAGF
Mvan_4054|M.vanbaalenii_PYR-1 MSVIATVDLS---------------RWRAGGAAADAVAADVDEGLQRAGF
TH_4524|M.thermoresistible__bu MAGDFTSVPVIDIS----------ALRHGSGPGYRRAVTALGDAARHVGF
*. : :..**
MSMEG_0414|M.smegmatis_MC2_155 FYLVGHGVPQDLVDRVLTAARRLFALPQADKDAIAMVNSPH-FRGYTRLG
MAV_4913|M.avium_104 FYLTGHGVPADLVDRVLDAARRFFRLPQADKDAVAMVRSPH-FRGYTRLG
MAB_4434c|M.abscessus_ATCC_199 FYLTGHGVSPQLVHEVLSAARKLFALSEADKEAIAMVRSPH-FRGFTRLG
Mflv_2580|M.gilvum_PYR-GCK ILVTGHGVDPDLARDVRAAAREFFALDPTVKAGYSVPVGGRGWIGPGAEA
Mvan_4054|M.vanbaalenii_PYR-1 ILVSGHGVSASLARDVRAAAREFFALPDEVKSGYSVPVGGHGWIGPGAEA
TH_4524|M.thermoresistible__bu AQLVGHGIDPALFDALVDRTRAFFALPAAEKHRVYIGNSTN-HRGYVPPG
: ***: * : :* :* * * : . . * .
MSMEG_0414|M.smegmatis_MC2_155 GELTGGQVDWREQIDIGPEREPLDDPAEP-YLRLQGPNQWPAALPELPAV
MAV_4913|M.avium_104 GERTRGAVDWREQIDIGPQRPPIGGLDKSDYRWLQGPNQWPAALPELPGL
MAB_4434c|M.abscessus_ATCC_199 GELTGGLTDWREQIDVGPERAASPTADGRDYMWLQGPNQWPAAVPELRPA
Mflv_2580|M.gilvum_PYR-GCK NGYAEGTETPPDLKESFSLGAETATGDAEVDRIWFAPNVWPREVPELAPL
Mvan_4054|M.vanbaalenii_PYR-1 NGYAEGTETPPDLKETFSLGAETATGDAEVDRIWFAPNVWPREVPALQPL
TH_4524|M.thermoresistible__bu EEVFAGGTADTKEAFDLSYELPADDPRYLAGDPLLGPNQWP-ELTGFREA
* . . .** ** :. :
MSMEG_0414|M.smegmatis_MC2_155 IAEWDAALAQVGRALLRHWAVALGNPADVFDEAFADTPATLIKIVRYPAQ
MAV_4913|M.avium_104 IAEWDAALSGVARTLLRHWAAALGSPADVFDAAFADAPATLIKIIRYPGR
MAB_4434c|M.abscessus_ATCC_199 LESWDIALAEVGRKLLREWAIALGSDGAVFSEAFGDKPATLIKVIRYPSD
Mflv_2580|M.gilvum_PYR-GCK VEEYTAQMRRVADDLLALCAHALGLSVNPFAALAS-APTWTMNINHYPPV
Mvan_4054|M.vanbaalenii_PYR-1 LTEYTAQMRGVADDLLALFAHALKLPANPFAELAS-TPTWTMNINHYPPV
TH_4524|M.thermoresistible__bu VTAWYDAVFALSRAILRAFAVALGQPPDRFDRYVT-TPPSQLRLIHYPAD
: : : :. :* * ** * *. :.: :**
MSMEG_0414|M.smegmatis_MC2_155 AETSQ------GVGAHRDAGVLTLLLAEPGSRGLQVR--GPDG-VFIDVD
MAV_4913|M.avium_104 AAGPQ------GVGAHRDAGVLTLLLAEPGSRGLQVRRGGPRGDGWIDVP
MAB_4434c|M.abscessus_ATCC_199 GTTSQ------GVGAHRDSGVLTLLLTEPEVGGLQVE---VDG-AWLEAP
Mflv_2580|M.gilvum_PYR-GCK SVVGEPEPGQFRIGPHTDFGTVTILDREPGAGGLQVY---SEQAGWENAP
Mvan_4054|M.vanbaalenii_PYR-1 SVVGEPEPGQFRIGPHTDFGTVTILDREPGAGGLQVY---SEDAGWQDAP
TH_4524|M.thermoresistible__bu PSAVDRP----GIGAHTDYECFTLLR--PTAPGLEVLN---AAGQWIDVP
: :*.* * .*:* * **:* : :.
MSMEG_0414|M.smegmatis_MC2_155 PLPGAFIVNIGEMLEIASGGYLRATEHRVQVSAS-----ERISVPYFFNP
MAV_4913|M.avium_104 PRQGAFIVNIGELLEAATRGYLRATEHRVNLAGSTA---ERISVPYFFNP
MAB_4434c|M.abscessus_ATCC_199 GREGAFIVNIGELLERASGGYLRATRHRVTLTGAP-----RISVAYFFNP
Mflv_2580|M.gilvum_PYR-GCK YDPAALTVNIGDLLEYWSGRRWPSGRHRVLPPQPDAPEEDLVSLIYFYEA
Mvan_4054|M.vanbaalenii_PYR-1 YDPDALTVNIGDLLEYWSGRRWPSGRHRVLPPQPGAPEEDLVSLIYFYEA
TH_4524|M.thermoresistible__bu YLDDAFVLNIGDLLEIWTNGAFVATTHRVRRVTS-----ERYSFPLFVNV
*: :***::** : : *** . *. * :
MSMEG_0414|M.smegmatis_MC2_155 RLDAAIPVLALPDELAAQA---RGVTADPSNDRIYSVYGRNAWKSRVRAH
MAV_4913|M.avium_104 RLDARIPVLSLPPELRARAG-ERWTPSEDPADPIFSVYGRNAWKSRLRSH
MAB_4434c|M.abscessus_ATCC_199 RLDARIPVLELPPELRDRA---RGVSAD-PDDPIHATYGENAWKSRLRAH
Mflv_2580|M.gilvum_PYR-GCK NHDALVTPLSPPIGR---------------AEGLEPVLSADFIKQRLDAI
Mvan_4054|M.vanbaalenii_PYR-1 NHDALVTPLDPPVGR---------------VSGLAPVTSADFIKERLDAI
TH_4524|M.thermoresistible__bu DADTEVAPLAEFVDGTARYAPVIAGEHLFAQTARSFRYLQDRIRRGEVSL
*: :. * : : :
MSMEG_0414|M.smegmatis_MC2_155 PDVAAAHGYV--------
MAV_4913|M.avium_104 PDVASAHGFSAHHGAAD-
MAB_4434c|M.abscessus_ATCC_199 PDVAAVHGHLDS------
Mflv_2580|M.gilvum_PYR-GCK TVS---------------
Mvan_4054|M.vanbaalenii_PYR-1 TVG---------------
TH_4524|M.thermoresistible__bu PDGAYPLSAFGRRAASPR
.