For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADTQTTQPSGLKNMARAVVGALPFVTRSESLPKRTVTVDDLTIDPANVAEYAQVTGLRFGDAVPLTYPFT LTFPAVMSLVTGLDFPFAAMGSVHIENHITQYRPIAVTDTVGVAVHAENLREHRRGLLVDIVTDVKVGNE LAWHQVTTFLHQQRTSLSGEPKPPPQKQPKLPPPNAVLRITPGQIRHYAAVGGDHNPIHTNAIAAKLFGF PTVIAHGMFSAAAVLANIEGQLSDAVKHSVRFAKPVVLPAKAALYVERDADGWELTLRHLTKGYPHLTAT VTPL*
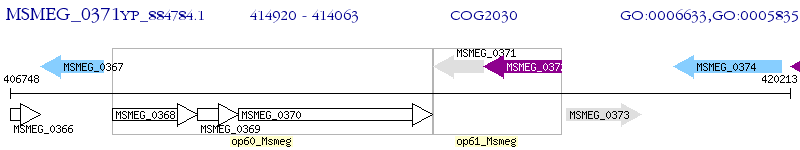
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0371 | - | - | 100% (285) | MaoC like domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6754 | - | 2e-09 | 37.04% (81) | MaoC like domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1341 | - | 3e-07 | 35.44% (79) | (3R)-hydroxyacyl-ACP dehydratase subunit HadB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0247c | - | 1e-129 | 78.85% (279) | hypothetical protein Mb0247c |
| M. gilvum PYR-GCK | Mflv_0413 | - | 1e-136 | 82.01% (278) | dehydratase |
| M. tuberculosis H37Rv | Rv0241c | - | 1e-129 | 78.85% (279) | hypothetical protein Rv0241c |
| M. leprae Br4923 | MLBr_02566 | - | 1e-123 | 74.82% (282) | hypothetical protein MLBr_02566 |
| M. abscessus ATCC 19977 | MAB_4444 | - | 1e-102 | 61.57% (281) | hypothetical protein MAB_4444 |
| M. marinum M | MMAR_0502 | - | 1e-128 | 78.65% (281) | hypothetical protein MMAR_0502 |
| M. avium 104 | MAV_4917 | - | 1e-125 | 77.70% (278) | MaoC like domain-containing protein |
| M. thermoresistible (build 8) | TH_1913 | - | 1e-120 | 75.65% (271) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1165 | - | 1e-127 | 78.29% (281) | hypothetical protein MUL_1165 |
| M. vanbaalenii PYR-1 | Mvan_0258 | - | 1e-140 | 84.34% (281) | dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0413|M.gilvum_PYR-GCK ---------------MSD---KQPSGLKNLALAAAGALPFVPRGDTLPTR
Mvan_0258|M.vanbaalenii_PYR-1 ---------------MNES--KQPSGLKNLALAVAGALPLVPRGDTLPTR
MSMEG_0371|M.smegmatis_MC2_155 ---------------MADTQTTQPSGLKNMARAVVGALPFVTRSESLPKR
MMAR_0502|M.marinum_M --------------------MNQPSGLKNMLRAAAGALPVVPRGDALPNR
MUL_1165|M.ulcerans_Agy99 --------------------MNQPSGLKNMLRAAAGALPVVPRGDALPNR
Mb0247c|M.bovis_AF2122/97 --------------------MTQPSGLKNLLRAAAGALPVVPRTDQLPNR
Rv0241c|M.tuberculosis_H37Rv --------------------MTQPSGLKNLLRAAAGALPVVPRTDQLPNR
MLBr_02566|M.leprae_Br4923 MTFGFVLRRLRRHFSVKENTVNQPSGLKNILRAIVGALPLLPRTDQLPSR
MAV_4917|M.avium_104 -------------------MMNQPSGLKNMLCAAAGALPVVSRGSALPTR
TH_1913|M.thermoresistible__bu -----------------------------MVRAAAGALPVVRRDGTLPKR
MAB_4444|M.abscessus_ATCC_1997 --------------------MAQPSGLQNMVMAAVGALPFIPRPSTLPTR
: * .****.: * **.*
Mflv_0413|M.gilvum_PYR-GCK TLTVEDLSIDPVNVAEYADVTGLRFDNTVPLTYPFALTFPTVMQLITGFD
Mvan_0258|M.vanbaalenii_PYR-1 TLTVEDLTIDPVNVAEYANVTGLRFGNTVPLTYPFALTFPTVMALLTGFD
MSMEG_0371|M.smegmatis_MC2_155 TVTVDDLTIDPANVAEYAQVTGLRFGDAVPLTYPFTLTFPAVMSLVTGLD
MMAR_0502|M.marinum_M TVSVEELPIDHTNVAAYAAVTGLRYRNNVPLTYPFVLAFPSVMALVTGFD
MUL_1165|M.ulcerans_Agy99 TVSVEELPIDHTNVAAYAAVTGLRYRNNVPLTYPFVLAFPSVMALVTGFD
Mb0247c|M.bovis_AF2122/97 TVTVEELPIDPANVAAYAAVTGLRYGNQVPLTYPFALTFPSVMSLVTGFD
Rv0241c|M.tuberculosis_H37Rv TVTVEELPIDPANVAAYAAVTGLRYGNQVPLTYPFALTFPSVMSLVTGFD
MLBr_02566|M.leprae_Br4923 TVTIEELPIDHTNVSAYASVTGLRYGNHVPLTYPFALTFPAMMSLVTGFD
MAV_4917|M.avium_104 TVTVEDLPIDRTNVAEYAAVTGLRYRNHVPLTYPFALTFPALMSLVTGFD
TH_1913|M.thermoresistible__bu TVTATDLPIDAAHVADYAAVTGLLFRDQVPLTYPFALTFPHVMELLSGFD
MAB_4444|M.abscessus_ATCC_1997 VVRTQGVKIDPANVAAYAQVTGLTFSDKVPVTYPFTLQFPSVMSLVASLD
.: : ** .:*: ** **** : : **:****.* ** :* *::.:*
Mflv_0413|M.gilvum_PYR-GCK FPFAAMGSVHLENHITQLRPIAVTDTVSVAVHAENMREHRRGLLVDILTD
Mvan_0258|M.vanbaalenii_PYR-1 FPFAAMGSVHLENHITQYRPIAVTDTVSVGVHAENMREHRRGLLVDVVTD
MSMEG_0371|M.smegmatis_MC2_155 FPFAAMGSVHIENHITQYRPIAVTDTVGVAVHAENLREHRRGLLVDIVTD
MMAR_0502|M.marinum_M FPFAAMGSVHTENHITQYRPIAVTDTVGVKVHAENLREHRKGLMVDLVTD
MUL_1165|M.ulcerans_Agy99 FPFAAMGSVHTENHITQYRPIAVTDTVGVKVHAENLREHRKGLMVDLVTD
Mb0247c|M.bovis_AF2122/97 FPFAAMGAIHTENHITQYRPIAVTDAVGVRVRAENLREHRRGLLVDLVTN
Rv0241c|M.tuberculosis_H37Rv FPFAAMGAIHTENHITQYRPIAVTDAVGVRVRAENLREHRRGLLVDLVTN
MLBr_02566|M.leprae_Br4923 FPFAAMGSVHTENHITQYRPIAVTDVVGVQVHAENLREHRKGLLVDLVTD
MAV_4917|M.avium_104 FPFAAMGSVHTENHITQYRPIAVTDTVGVRVHAENLREHRKGLLVDLVTD
TH_1913|M.thermoresistible__bu FPFPAIGAVHVENHITQQRPILVTDIVNISVHAENLREHRRGLLVDLVTD
MAB_4444|M.abscessus_ATCC_1997 FPFPAIGTVHLENHITQHRAISATDTVDIEVRAENLREHRKGLLVDVLTD
***.*:*::* ****** *.* .** *.: *:***:****:**:**::*:
Mflv_0413|M.gilvum_PYR-GCK VRVGNELAWQQVTTFLHQQRTSLSDEPKPPPQKQPKLPPPNAVLRITPAQ
Mvan_0258|M.vanbaalenii_PYR-1 VKVGNDPAWHQITTFLHQQRTSLSDQPKPPPQKQPKLPPPNAVLRITPAQ
MSMEG_0371|M.smegmatis_MC2_155 VKVGNELAWHQVTTFLHQQRTSLSGEPKPPPQKQPKLPPPNAVLRITPGQ
MMAR_0502|M.marinum_M INVGNELAWHQVTTFLHQQRTSLSGEPKPPPQKQPKLPPPAAVLRITPAQ
MUL_1165|M.ulcerans_Agy99 INVGNELAWHQVTTFLHQQRTSLSGEPKPPPQKQPKLPPPAAVLRITPAQ
Mb0247c|M.bovis_AF2122/97 VSVGNDVAWHQVTTFLHQQRTSLSGEPKPPPQKKPKLPPPAAVLRITPAK
Rv0241c|M.tuberculosis_H37Rv VSVGNDVAWHQVTTFLHQQRTSLSGEPKPPPQKKPKLPPPAAVLRITPAK
MLBr_02566|M.leprae_Br4923 VSVGNDTAWHQVTTFLHLQRTSLSDEPKPPSQKQPKLLPPSAVLQITPRQ
MAV_4917|M.avium_104 VSVGNDTAWHQVTTFLHQQRTSLSDQPKPPPQKQPKLPPPPTVLRITPGL
TH_1913|M.thermoresistible__bu IYVGNEAAWHQVSTFLHQQRTSLSDEPKPPPAKQPKLGPPNAVLRVTPGQ
MAB_4444|M.abscessus_ATCC_1997 ITIGTDTVWQQTMTFLRQQRTSLSGGPKPEPPKAVKLPPPNSTLRISGGQ
: :*.: .*:* ***: ******. *** . * ** ** :.*:::
Mflv_0413|M.gilvum_PYR-GCK IKHYASVGGDHNPIHTNGIAARLFGFPTVIAHGMFTAAAVLANVEGQLPD
Mvan_0258|M.vanbaalenii_PYR-1 IRHYASVGGDHNPIHTNAIAAKLFGFPTVIAHGMFSAAAVLANIEGQLPD
MSMEG_0371|M.smegmatis_MC2_155 IRHYAAVGGDHNPIHTNAIAAKLFGFPTVIAHGMFSAAAVLANIEGQLSD
MMAR_0502|M.marinum_M IRRYAAVGGDHNPIHTNSIAAKLFGFPTVIAHGMFSAAAVLANIEAKIPD
MUL_1165|M.ulcerans_Agy99 IRRYAAVGGDHNPIHTNSIAAKLFGFPTVIAHGMFSAAAVLANIEAKIPD
Mb0247c|M.bovis_AF2122/97 IRRYAAVGGDHNPIHTNPIAAKLFGFPTVIAHGMFTAAAVLANIEARFPD
Rv0241c|M.tuberculosis_H37Rv IRRYAAVGGDHNPIHTNPIAAKLFGFPTVIAHGMFTAAAVLANIEARFPD
MLBr_02566|M.leprae_Br4923 IRRYAAVGGDHNPIHTNPIVAKLFGFPTTIAHGMFSAAAVLANIEARLPD
MAV_4917|M.avium_104 IRRYAVVGGDHNPIHTNPIAAKLFGFPTVIAHGMFSAAAVLANIEAAIPD
TH_1913|M.thermoresistible__bu IRRYASVGGDHNPIHTSSLGAKLFGFPTVIAHGMYTAAAVLANIEGRLPD
MAB_4444|M.abscessus_ATCC_1997 IRRYASVGGDHNPIHTSSIGAKALGFPKAIAHGMFTAAAVLGNIQGDIPD
*::** **********. : *: :***..*****::*****.*::. :.*
Mflv_0413|M.gilvum_PYR-GCK GVKYSVRFAKPVILPARAGLYVDRRAD---GWDLTLRHLTKGHPHLTGTL
Mvan_0258|M.vanbaalenii_PYR-1 AVKYSVRFAKPVVLPARAGLYVDRSAD---AWELTLRHLTKGYPHLQATV
MSMEG_0371|M.smegmatis_MC2_155 AVKHSVRFAKPVVLPAKAALYVERDAD---GWELTLRHLTKGYPHLTATV
MMAR_0502|M.marinum_M AVRYSVRFGKPVLLPATAGLYIDEADAADPGWEIALRNMSKGYPHLTATI
MUL_1165|M.ulcerans_Agy99 AVRYSVRLGKPVLLPATAGLYIDEADAADPGWEIALRNMSKGYPHLTATI
Mb0247c|M.bovis_AF2122/97 AVRYSVRFAKPVLLPATAGLYVAEGDG---GWDLTLRNMAKGYPHLTATV
Rv0241c|M.tuberculosis_H37Rv AVRYSVRFAKPVLLPATAGLYVAEGDG---GWDLTLRNMAKGYPHLTATV
MLBr_02566|M.leprae_Br4923 AVHYSARFVKPVVLPATTGLYVDESAG---NWDLTLRNIAKGYPHLAGTV
MAV_4917|M.avium_104 AVRYSVRFGKPVVLPATTGLYIDEHDG---GWDIALRNIAKGYPHLTGTV
TH_1913|M.thermoresistible__bu AVRYSVRFGKPLVLPAAAGLYVQQTDG---GWELALRHLTKGYPHLTAMV
MAB_4444|M.abscessus_ATCC_1997 AVRYDVKFGKPVLLPAAVGVWIDRPGGPGTAWDIAVRNPRNGDPHVTGSV
.*::..:: **::*** ..::: . *::::*: :* **: . :
Mflv_0413|M.gilvum_PYR-GCK TSL-
Mvan_0258|M.vanbaalenii_PYR-1 TSLR
MSMEG_0371|M.smegmatis_MC2_155 TPL-
MMAR_0502|M.marinum_M KPL-
MUL_1165|M.ulcerans_Agy99 KPL-
Mb0247c|M.bovis_AF2122/97 RGL-
Rv0241c|M.tuberculosis_H37Rv RGL-
MLBr_02566|M.leprae_Br4923 QGV-
MAV_4917|M.avium_104 RAL-
TH_1913|M.thermoresistible__bu TPL-
MAB_4444|M.abscessus_ATCC_1997 TPL-
: