For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSNRLDRVRAAWDGIGVQTQFYARAIGSIGDAAMNYRTELIRLIATMGLGAGALAVVGGTVAIVGFLTMT TGALVAVQGYNQFASVGVEALTGFASAFFNVRLIAPGTTAVALSATIGAGATAQLGAMRINEEIDALEVI GIRSVSYLASTRVAAGVIVVIPLYCVAVIMSFISSRLGTTVIYGQGSGVYDHYFNTFLNTNDLLWSFGQS VAITVVIMLVHTYYGYTASGGPAGVGEAVGRAVRTSLIVAAIVVVMISLALYGQSGNFHLAG
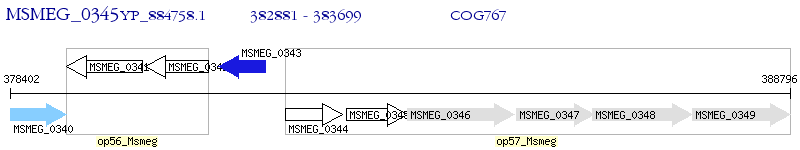
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0345 | - | - | 100% (272) | hypothetical protein MSMEG_0345 |
| M. smegmatis MC2 155 | MSMEG_4795 | - | 3e-89 | 56.62% (272) | ABC-transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_2854 | - | 2e-88 | 59.30% (258) | hypothetical protein MSMEG_2854 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3530c | yrbE4B | 3e-87 | 55.64% (266) | integral membrane protein YrbE4b |
| M. gilvum PYR-GCK | Mflv_2100 | - | 1e-113 | 74.72% (269) | hypothetical protein Mflv_2100 |
| M. tuberculosis H37Rv | Rv1965 | yrbE3B | 1e-104 | 69.62% (260) | integral membrane protein YrbE3b |
| M. leprae Br4923 | MLBr_02588 | yrbE1B | 1e-83 | 55.81% (258) | hypothetical protein MLBr_02588 |
| M. abscessus ATCC 19977 | MAB_4154c | - | 3e-85 | 56.49% (262) | hypothetical protein MAB_4154c |
| M. marinum M | MMAR_4050 | yrbE3B_1 | 1e-111 | 73.33% (270) | integral membrane protein YrbE3B_1 |
| M. avium 104 | MAV_2060 | - | 1e-101 | 68.22% (258) | TrnB2 protein |
| M. thermoresistible (build 8) | TH_4199 | - | 1e-120 | 82.06% (262) | PUTATIVE protein of unknown function DUF140 |
| M. ulcerans Agy99 | MUL_3917 | yrbE3B_1 | 1e-111 | 73.33% (270) | integral membrane protein YrbE3B_1 |
| M. vanbaalenii PYR-1 | Mvan_0362 | - | 1e-121 | 81.51% (265) | hypothetical protein Mvan_0362 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3530c|M.bovis_AF2122/97 ---MS------YDVTIRFRRFFSRLQRPVDNFGEQALFYGETMRYVPNAI
MAB_4154c|M.abscessus_ATCC_199 ---MSGPYLSTHTVTSYTVRYMRGLGRSFDKFGEQALFYAQSLSYIPAAL
MSMEG_0345|M.smegmatis_MC2_155 ----------MSNRLDRVR-------AAWDGIGVQTQFYARAIGSIGDAA
TH_4199|M.thermoresistible__bu -MAGVVTVGGPSRPLPRLRNVWGGLVRAWNRIGRQTEFYWRTVVAVPDAL
Mvan_0362|M.vanbaalenii_PYR-1 -----MTQSAPSRPHPRWYRAAHEVAAGWNELGDQARFYAGTLRAVPDAV
Rv1965|M.tuberculosis_H37Rv ----------MTAAKALVS--------EWNRMGSQMRFFVGTLAGIPDAL
MMAR_4050|M.marinum_M -----MTAVAAGSRWRGVRHKLDGWVAGWNQIGTQTKFFGRTLRSIHYML
MUL_3917|M.ulcerans_Agy99 -----MTAVAAGSRWRGVRHKLDGWVAGWNQIGTQTKFFGRTLRSIHYML
Mflv_2100|M.gilvum_PYR-GCK -----MTVSRPHSQFPWLKARFRSIADPWNQVGVQTKFYGRTLRSIVIAV
MAV_2060|M.avium_104 -------MTEIAREPSRFAALGPRLVESTRKLGEQTVFYGQSLTAIADAI
MLBr_02588|M.leprae_Br4923 MSTAAVLRARFPRAAANLNRYGGAAARGIDDIGQMSWFGLIALGNIPNAL
.* * :: :
Mb3530c|M.bovis_AF2122/97 TRYRKETVRLVAEMTLGAGALVMIGGTVGVAAFLTLASGGVIAVQGYSSL
MAB_4154c|M.abscessus_ATCC_199 TKYRKETIRVLAEITMGTGALVMIGGTVGVAVFLTLASGGVIAVQGYSSL
MSMEG_0345|M.smegmatis_MC2_155 MNYRTELIRLIATMGLGAGALAVVGGTVAIVGFLTMTTGALVAVQGYNQF
TH_4199|M.thermoresistible__bu MHYRAELIRLIAGMGLGTGALAIVGGTVAIVGFLTITTGGLVAVQGYNQL
Mvan_0362|M.vanbaalenii_PYR-1 RRYPGELLRLIAQMGLGTGALAVVGGTVAIVGFLTITTGALVAVQGYNQF
Rv1965|M.tuberculosis_H37Rv MHYRGELLRVIAQMGLGTGVLAVIGGTVAIVGFLAMTTGAIVAVQGYNQF
MMAR_4050|M.marinum_M IHYRVELIRLIAQMGLGAGALMVIGGTVAIVGFLTVTTGALVAVQGYSDF
MUL_3917|M.ulcerans_Agy99 IHYRVELIRLIAQMGLGAGALMVIGGTVAIVGFLTVTTGALVAVQGYSDF
Mflv_2100|M.gilvum_PYR-GCK TRYRIELVRQIAQMGLGAGALIVIGGTVAIVGFLTVTTGALVAVQGYTDF
MAV_2060|M.avium_104 RRYPGELLRLIAEMGMGTGALAVIGGTVGIIGFLTLTTGALVAVQGYDTL
MLBr_02588|M.leprae_Br4923 SRYRKETLRLIAQIGMGTGAMAVVGGTAAIVGFVTLSGSSLVAIQGFASL
.* * :* :* : :*:*.: ::***..: *:::: ..::*:**: :
Mb3530c|M.bovis_AF2122/97 GDIGIEALTGFLSAFLNVRVVAPVIAGIALAATIGAGATAQLGAMRVSEE
MAB_4154c|M.abscessus_ATCC_199 GNIGIEALTGFLSAFLNVRIVAPVVAGIAMAATIGAGTTAQLGAMRVAEE
MSMEG_0345|M.smegmatis_MC2_155 ASVGVEALTGFASAFFNVRLIAPGTTAVALSATIGAGATAQLGAMRINEE
TH_4199|M.thermoresistible__bu ASVGFEALTGFASAFFNVRLIVPGTTAVALSATIGAGATAQIGAMRINEE
Mvan_0362|M.vanbaalenii_PYR-1 ASVGVEALTGFASAFFNVRLIVPGTTAVALSATIGAGATAQLGAMRINEE
Rv1965|M.tuberculosis_H37Rv ASVGVEALTGFASAFFNTREIQPGTVMVALAATVGAGTTAALGAMRINEE
MMAR_4050|M.marinum_M SELGVEALTGFASAFFNVRLIAPATTAIALSATIGAGATAQLGAMRINDE
MUL_3917|M.ulcerans_Agy99 SELGVEALTGFASAFFNVRLIAPATTAIALSATIGAGATAQLGAMRINDE
Mflv_2100|M.gilvum_PYR-GCK SEIGVEALTGFASAFFNVRLIAPATTAVALAATIGAGATAQLGAMRINEE
MAV_2060|M.avium_104 SNIGVEALTGFLSAFLNVRMIAPCTAGLALAATIGAGATAQLGAMRINEE
MLBr_02588|M.leprae_Br4923 GAIGVEAFTGFFAALINVRIAAPVITGIVMAATVGAGATAELGAMRISEE
. :*.**:*** :*::*.* * . :.::**:***:** :****: :*
Mb3530c|M.bovis_AF2122/97 IDAVECMAVHSVSYLVSTRLIAGLVAIIPLYSLSVLAAFFAARFTTVFVN
MAB_4154c|M.abscessus_ATCC_199 IDALETMAVHAISYLVSTRLVAGLIAIIPLYSLSVLAAFFASRATTVMIN
MSMEG_0345|M.smegmatis_MC2_155 IDALEVIGIRSVSYLASTRVAAGVIVVIPLYCVAVIMSFISSRLGTTVIY
TH_4199|M.thermoresistible__bu IDALEVIGIRSVTYLASTRVLAGVIVVVPLYCVAMIMAFLSARTGTTVIY
Mvan_0362|M.vanbaalenii_PYR-1 IDALEVIGIRSVTYLAATRVLAGVVVVIPLYCIAVMMAFLAARTGTTVIY
Rv1965|M.tuberculosis_H37Rv IDALEVIGIRSISYLASTRVLAGVVVAVPLFCVGLMTAYLAARVGTTAIY
MMAR_4050|M.marinum_M IDALEVMGIRTIAFLAASRVVAGLLVVIPLYCVGVLASFWAARFGTTVLY
MUL_3917|M.ulcerans_Agy99 IDALEVMGIRTIAFLAASRVVAGLLVVIPLYCVGVLASFWAARFGTTVLY
Mflv_2100|M.gilvum_PYR-GCK IDALEVMGIRSVAYLASTRVLAGLLVVIPLYCVGVLASFWAARFGTTVIY
MAV_2060|M.avium_104 IDALEVMGIRAVTYLASTRIIAGILVVIPLYAVAVLMSFIAAKLLTIYVY
MLBr_02588|M.leprae_Br4923 IDALEVMSIKSISFLVTTRILAGLVVIVPLYAVAMIMAFLSPHIITTVLY
***:* :.::::::*.::*: **::. :**:.:.:: :: :.: * :
Mb3530c|M.bovis_AF2122/97 GQSAGLYDHYFNTFLIPSDLLWSFMQAIAMSIAVMLVHTYYGYNASGGSV
MAB_4154c|M.abscessus_ATCC_199 GQSPGVYDHYFNTFLVPTDLLWSFLQAIVMSVVVMLVHTSYGFNASGGPV
MSMEG_0345|M.smegmatis_MC2_155 GQGSGVYDHYFNTFLNTNDLLWSFGQSVAITVVIMLVHTYYGYTASGGPA
TH_4199|M.thermoresistible__bu GQGSGVYDHYFNTFLNPTDLIWSFFQVVALAVVIMLVHTFYGYTASGGPA
Mvan_0362|M.vanbaalenii_PYR-1 GQGSGVYDHYFNTFLNPTDLVWSFLQSVAITVVVMLVHTYYGYTARGGPA
Rv1965|M.tuberculosis_H37Rv GQGSGVYDHYFNTFLRPTDVLWSSVEVVVVALMIMLVCTYYGYAAHGGPA
MMAR_4050|M.marinum_M SQATGVYDHYFRTFLNPTDLVWSFVQCIALSLVIMLVHTYYGYTARGGPA
MUL_3917|M.ulcerans_Agy99 SQATGVYDHYFRTFLNPTDLVWSFVQCIALSLVIMLVHTYYGYTARGGPA
Mflv_2100|M.gilvum_PYR-GCK GQSTGVYDHYFRTFLNPTDLVWSFAQCIVLAIVIMLVHTYYGFSARGGPA
MAV_2060|M.avium_104 GQSKGVYEHYFSTFLRPNDLLWSFFSALAMATGVMVVHTYYGFTATGGPA
MLBr_02588|M.leprae_Br4923 GQSNGTYEHYFRTFLRPDDVFWSFLEAVIITAVVMITHCYYGYTANGGPV
.*. * *:*** *** . *:.** . : :: :*:. **: * **..
Mb3530c|M.bovis_AF2122/97 GVGVAVGQAVRTSLIVVVVITLFISLAVYGASGNFNLSG
MAB_4154c|M.abscessus_ATCC_199 GVGVAVGQAVRTSLIVVVLVTLFISLAVYGGSGNFNLSG
MSMEG_0345|M.smegmatis_MC2_155 GVGEAVGRAVRTSLIVAAIVVVMISLALYGQSGNFHLAG
TH_4199|M.thermoresistible__bu GVGEAVGRAVRTSMIVAAVVIIFISLAAYGQTGNFVLAG
Mvan_0362|M.vanbaalenii_PYR-1 GVGEAVGRAVRTSMVVAAVEIVMISLAIYGQSGNFNLAG
Rv1965|M.tuberculosis_H37Rv GVGEAVGRAVRASMVVASIAILVMTLAIYGQSPNFHLAT
MMAR_4050|M.marinum_M GVGEAVGRAVRTSLIFSAFVLVMISLAVYGQSGNFNLAG
MUL_3917|M.ulcerans_Agy99 GVGEAVGRAVRTSLIFSAFVLVMISLAVYGQSGNFNLAG
Mflv_2100|M.gilvum_PYR-GCK GVGEAVGRAVRTSLILSAFVLVMISLAVYGQSGNFNLAG
MAV_2060|M.avium_104 GVGEAVGRSVRSSMIVTAFVCLMISLSVYGQSGNFNLSG
MLBr_02588|M.leprae_Br4923 GVGEAVGRSMRFSLVSVQVVVLSATLALYGVDPNFALTV
*** ***:::* *:: . : :*: ** ** *: