For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTIIGGGILGTAHAVEAVRRGHRVVHLEREAEARGATVRNFGLVWISGRSTAELDATLRSRELWEKLAVD VPGIGFRPAGSITLVRTPAELAVAQEAATRSDAGHRGFTLLEPDEVRELNPALRGRFLGGLHCALDAAVE SRQALPAIREHLTASGRYEFVAGVEARGVENRTVRDDRGNRHESDIVLVCAGAAHGGLVRDLAGELPVRR VRLQMMQTEPLGERLTTAIADADSFRYYPGFAGPALNTLRDNQPQHRVADGQHMQLLCVQRLHGGLTIGD THEYTEPFGFDVHEEPYTYLLDVVEEFLGRKLPPIRQRWAGVYSQCLDPEQLVCRTAPEDDIWVITGPGG RGMTLGPAIAEQTADMIGL
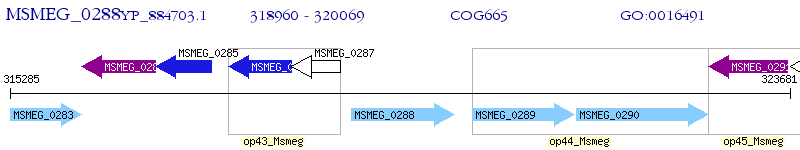
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0288 | - | - | 100% (369) | FAD dependent oxidoreductase, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3080 | - | 1e-09 | 28.69% (244) | FAD dependent oxidoreductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1499 | - | 1e-138 | 65.15% (373) | putative FAD dependent oxidoreductase |
| M. marinum M | MMAR_4421 | solA | 2e-05 | 24.68% (389) | dehydrogenase |
| M. avium 104 | MAV_0142 | - | 2e-05 | 27.04% (196) | FAD dependent oxidoreductase, putative |
| M. thermoresistible (build 8) | TH_0796 | - | 1e-79 | 70.20% (198) | FAD dependent oxidoreductase, putative |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0054 | - | 1e-157 | 72.63% (369) | FAD dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1499|M.abscessus_ATCC_1997 -----MRVIVVGGGILGTAHAWAAVERGHEVVHLEREV---EARGATVRN
Mvan_0054|M.vanbaalenii_PYR-1 -----MQVTIIGAGILGTAHAWHAVRRGHHVIHLEREV---EARGATVRN
MSMEG_0288|M.smegmatis_MC2_155 -------MTIIGGGILGTAHAVEAVRRGHRVVHLEREA---EARGATVRN
TH_0796|M.thermoresistible__bu -------VIIIGGGILGTAHAVEAVRRGHQVVQLEREG---EARGATVRN
MMAR_4421|M.marinum_M MATTGYDVIVIGLGGMGSAAAYHLAARGRRVLGLERHQ---PAHDKGSSH
Mflv_3080|M.gilvum_PYR-GCK -MPRTPRVVVIGAGIVGTSLADELTARGWTDVTVVDRGPLFATGGSTSHA
MAV_0142|M.avium_104 ------MIVVIGAGVCGLAAAHELSRRGHDDVVVLEKG-----QPFGEQS
: ::* * * : * ** : : .
MAB_1499|M.abscessus_ATCC_1997 FGLVWVSG---RSALELKIAQRSRDLWQGIAKLVPG-IGFRQMGSFTLLR
Mvan_0054|M.vanbaalenii_PYR-1 FGLVWVSG---RASGELEASLRSRALWEEIGAEVPA-IGFRPCGSLTLLR
MSMEG_0288|M.smegmatis_MC2_155 FGLVWISG---RSTAELDATLRSRELWEKLAVDVPG-IGFRPAGSITLVR
TH_0796|M.thermoresistible__bu FGLVWVSG---RAPAELDTALRARELWERLGAAIPG-IGFRPAGSLTLLK
MMAR_4421|M.marinum_M GGSRIIRQSYFEDPGYVPLLLRSYELWERLAIDAQS-DIYRLTG--GLFI
Mflv_3080|M.gilvum_PYR-GCK PGLVFQTN-----PSKTMTVFARYTVEKFAALQHPGGWAFNPVGGLEVAT
MAV_0142|M.avium_104 AGLARIFRIAHQREALCRLAFAARAGWQRWEAEFGV---GRLLGSEGFIA
* : . * .
MAB_1499|M.abscessus_ATCC_1997 THEEVAVAEEVMARSDADERGFALLEPGAVQRQNPALRG-KFLAGLHCAR
Mvan_0054|M.vanbaalenii_PYR-1 TPAEVAVAEEVCGRADAASRGFRLLEPVEAREINPVLRG-KFLAALHCDR
MSMEG_0288|M.smegmatis_MC2_155 TPAELAVAQEAATRSDAGHRGFTLLEPDEVRELNPALRG-RFLGGLHCAL
TH_0796|M.thermoresistible__bu TLDEVAVAEQAAARADAGRRGFTVVDAEQARRLNPALRG-DFLAALHCSR
MMAR_4421|M.marinum_M GPPDCATVAGSLRASREWDLPHEVLDAPQIAARFPNFTP-QPTDIALYEA
Mflv_3080|M.gilvum_PYR-GCK TPERMADLHRKAGWAQSWGIDGRLLTPSECVQLHPLVDRDRILGGFHTPA
MAV_0142|M.avium_104 AG---AAADGVAQAMKRAGAAFSRLDRDGIAERIPFADV-PWETGIFDPL
* : *
MAB_1499|M.abscessus_ATCC_1997 DAAVESRVALPAMRLHLKTNGCYSFVAGREVRAIDTSSAGVSVRTDDG-E
Mvan_0054|M.vanbaalenii_PYR-1 DGAVESRQALPAIRAVLDATGRYTFVPGTEARAVD----GTRVHDDRG-N
MSMEG_0288|M.smegmatis_MC2_155 DAAVESRQALPAIREHLTASGRYEFVAGVEARGVEN----RTVRDDRG-N
TH_0796|M.thermoresistible__bu DAAVESRRAMPAIRAHLAGTGRYTFRPGTEARSVS----GPSVRDDRG-G
MMAR_4421|M.marinum_M KAGLAR--PELTVRAHLELAKRSGATLQFEEPVLNWTTTASGVRVTTGRD
Mflv_3080|M.gilvum_PYR-GCK DGLAKALRAAEAQARRAEARGATLRPHTEVLGILDDGRRVTGIRTADG--
MAV_0142|M.avium_104 GGSLRIRRALTALARRVVIRCGEVVSVADDG----------RTVLADG-T
. . : *
MAB_1499|M.abscessus_ATCC_1997 TYRGEAVVMCAGAAHGGLARELLGDFPVRRVRLQMMQTAALGEELTTAIA
Mvan_0054|M.vanbaalenii_PYR-1 TYDADVVIVCAGAAHGGLVRDLAGDLPVRRVRLQMMQTAPLGEQLTTAIA
MSMEG_0288|M.smegmatis_MC2_155 RHESDIVLVCAGAAHGGLVRDLAGELPVRRVRLQMMQTEPLGERLTTAIA
TH_0796|M.thermoresistible__bu RHDGDVVIVCAGAAHGGLX-------------------------------
MMAR_4421|M.marinum_M TYTAEQLVVCPGAWAPELLSQLGIPITVQRQVMYWLDP----------VG
Mflv_3080|M.gilvum_PYR-GCK TLDADIVVCAAGFWGAQLARQVGLVLPLVPMAHQYARTG-----QIAELV
MAV_0142|M.avium_104 VLRADRVLICAGVATPKLFGPLG---------------------------
.: :: ..* *
MAB_1499|M.abscessus_ATCC_1997 DGDSLRYYPAFAGPALDALNATQLQEPTAAEHKMQLLCVQRLHGGLTIGD
Mvan_0054|M.vanbaalenii_PYR-1 DGDSLRYYPGFAGGALDTLRRSESQDPVADEHHMQLLCVQRLHGGLTIGD
MSMEG_0288|M.smegmatis_MC2_155 DADSFRYYPGFAGPALNTLRDNQPQHRVADGQHMQLLCVQRLHGGLTIGD
TH_0796|M.thermoresistible__bu -GCGCR--------------------------------------------
MMAR_4421|M.marinum_M GTAGFRDHPIFIAENDSGAQIYGFPAIDGPRGGVKVAFFR--------KG
Mflv_3080|M.gilvum_PYR-GCK GRNTEIAEAGLPILRHQDQDLYFREHGDRLGIGSYMHRPMPVDMTTLLTD
MAV_0142|M.avium_104 --VGFVPHTRFTYRGADATGAACLSAPEGYGLPLGG------------TG
MAB_1499|M.abscessus_ATCC_1997 THEYAEPFAFDVDEAPYRHLITTAEELLGRKLPP-VVRRWAGVYSQCTDP
Mvan_0054|M.vanbaalenii_PYR-1 THEYTEPFDFDVTEAPYSYLTGVVEELLGRSLPP-IRRRWAGVYSQCVDP
MSMEG_0288|M.smegmatis_MC2_155 THEYTEPFGFDVHEEPYTYLLDVVEEFLGRKLPP-IRQRWAGVYSQCLDP
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M IACTPDTIDRVVGPQEIHEMITRVGELLPALAGT-CLQAATCMYSNTPDQ
Mflv_3080|M.gilvum_PYR-GCK TAGEAMPSMLPFTEEDFAPAWHAATDLIPALRDSKVEEAFNGIFSFTTDG
MAV_0142|M.avium_104 RWAFGQEVADPATVRALFPSLVAVDRVDCVSVRAPWLDSGGDGWTVARRG
MAB_1499|M.abscessus_ATCC_1997 DQIVYRRQAAPAVWVVTGPGGRG------------MTLGPAIGADTADLM
Mvan_0054|M.vanbaalenii_PYR-1 ERLAYREQVSPGVWVVTGPGGRG------------MTLGPVIGEQTTNEI
MSMEG_0288|M.smegmatis_MC2_155 EQLVCRTAPEDDIWVITGPGGRG------------MTLGPAIAEQTADMI
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M HFVIARHPYCPAVTVACGFSGHG------------FKFVPVVGEILADLV
Mflv_3080|M.gilvum_PYR-GCK FSIMGEHRDLAGFWVAEAVWVTHSAGVARATAEWIIEGTPTIDTHECDLY
MAV_0142|M.avium_104 RVLAFVGANLMKFGPLLGELLAR---------------AALSDDIPAELL
MAB_1499|M.abscessus_ATCC_1997 NL------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 GL------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 GL------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M TEGATSQPISLFDPQRLVTV------------------------------
Mflv_3080|M.gilvum_PYR-GCK RFEDVARSPRFIAQTSAQAFVEVYDVIHPHQYRTALRGLRTSPFYPRQCE
MAV_0142|M.avium_104 LD------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK LGASFYEGGGWERPAWYGANDRLVQRLFDDGLAVPHRDEWAARNWSPISI
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK AEAHWTRECVAMYDMTPLTRYEVSGSGAAAFLQQMTTNNVDKSVGSVTYT
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK LLLDESGGIRSDLTVARLGPEHFQVGANSPMDFDWLSRRKPPGVVVRDIT
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK GGTCCLGVWGPRAREVIAPLCPDNLSHKAFAYFRAMHTHLGAIPVVMMRV
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK SYVGELGWEIYAGAEYGAALWDLIDEAGAAHGIIPAGRIAFNSLRIEKGY
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK RSWGTDMTAEHRPAAAGLDFAVRVDKGDFVGRQALLTAGSPDATLRSIVF
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0054|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0288|M.smegmatis_MC2_155 --------------------------------------------------
TH_0796|M.thermoresistible__bu --------------------------------------------------
MMAR_4421|M.marinum_M --------------------------------------------------
Mflv_3080|M.gilvum_PYR-GCK DDPDAAVLGKEPVSVGDTCVGYVTSAGYSATIGRTIAYAWLPAGLVIGET
MAV_0142|M.avium_104 --------------------------------------------------
MAB_1499|M.abscessus_ATCC_1997 -----------------------------
Mvan_0054|M.vanbaalenii_PYR-1 -----------------------------
MSMEG_0288|M.smegmatis_MC2_155 -----------------------------
TH_0796|M.thermoresistible__bu -----------------------------
MMAR_4421|M.marinum_M -----------------------------
Mflv_3080|M.gilvum_PYR-GCK VSVAYRGVTYSATVHAEPVVDPDMSRIKR
MAV_0142|M.avium_104 -----------------------------