For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YDYGTFAFASKADVLDQAKAFWNPDKTQFWTDSGVDLVIDRREGYFLWDMSGRRLIDLHLNGGTYNIGHR NPEVMQAISEGMAHFDVGNHHFPSVARTALAKRLVETAPASIKKVAFGSGGGEAIDIALKSARHGTQRRK IVSVIKAYHGHTGLAVATGDDRFAKLFLADRPDEFIQVPFGDIGAMEAALAGGDVAAVIMETIPATYGFP LPPPGYLEAVKALTERHGTLYIADEVQTGLMRTGELWGITKHGIDPDIIVTGKGLSGGMYPISAVLLGER AARWLDQDGFGHISTFGGAELGCVAAIKTLEICTRPEVRSMVHYIADIFDTGLRRIQADYPDWFVGIRQN GVVIGLEFDHPEGAKFVMRELYENGVWAIFSTLDPRVLQFKPGLLLSRELCEDVLDRLEVAVGRARATVT GRRNG*
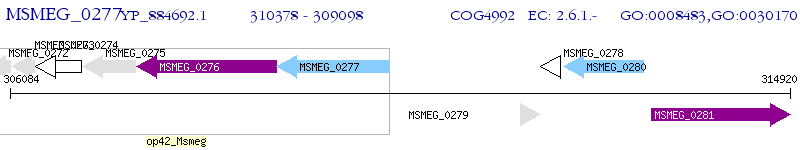
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0277 | - | - | 100% (426) | putative aminotransferase |
| M. smegmatis MC2 155 | MSMEG_2450 | - | 2e-38 | 28.57% (357) | hypothetical protein MSMEG_2450 |
| M. smegmatis MC2 155 | MSMEG_3773 | argD | 2e-32 | 30.21% (374) | acetylornithine aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1683 | argD | 6e-34 | 29.87% (375) | acetylornithine aminotransferase |
| M. gilvum PYR-GCK | Mflv_1243 | - | 0.0 | 84.67% (424) | aminotransferase class-III |
| M. tuberculosis H37Rv | Rv1655 | argD | 6e-34 | 29.87% (375) | acetylornithine aminotransferase |
| M. leprae Br4923 | MLBr_01409 | argD | 2e-34 | 29.14% (374) | acetylornithine aminotransferase |
| M. abscessus ATCC 19977 | MAB_2279 | - | 6e-43 | 30.87% (392) | ornithine aminotransferase |
| M. marinum M | MMAR_2465 | argD | 1e-35 | 31.05% (380) | acetylornithine aminotransferase ArgD |
| M. avium 104 | MAV_3115 | argD | 1e-34 | 29.92% (391) | acetylornithine aminotransferase |
| M. thermoresistible (build 8) | TH_3529 | - | 4e-35 | 30.77% (377) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_1647 | argD | 1e-36 | 31.32% (380) | acetylornithine aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_5565 | - | 0.0 | 85.14% (424) | aminotransferase class-III |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1683|M.bovis_AF2122/97 --------MTGASTTTATMRQRWQAVMMNNYGTP-----PIALASGDGAV
Rv1655|M.tuberculosis_H37Rv --------MTGASTTTATMRQRWQAVMMNNYGTP-----PIALASGDGAV
MLBr_01409|M.leprae_Br4923 --------MTPTQTNTATMQQRWETVMMNNYGTP-----PIVLASGNGAV
MAV_3115|M.avium_104 -----------MVTNTDALRRRWEAVMMHNYGTP-----PVALASGEGAV
MMAR_2465|M.marinum_M ------------MTHTEDTRARWEAVMMNNYGTP-----PVALASGEGAV
MUL_1647|M.ulcerans_Agy99 ------------MTHTEDTRARWEAVMMNNYGTP-----PVALASGEGAV
TH_3529|M.thermoresistible__bu ---------VSGSKGRHTLLDRWQGVMMNNYGTP-----PLALVSGDGAV
MAB_2279|M.abscessus_ATCC_1997 ----MSVTASPQAPSAAHIARIYKTHLSGGRATLGDIFGGHIETSSDGAW
Mflv_1243|M.gilvum_PYR-GCK MYDYGTFSFDSKAQVLERAKEFWNPDKTQFWTDTG---VDLVIDRRQDYF
Mvan_5565|M.vanbaalenii_PYR-1 MYDYGTFSFDSKAQVLERAKEFWNPDKTQFWTDSG---VDLVIDRRQDYF
MSMEG_0277|M.smegmatis_MC2_155 MYDYGTFAFASKADVLDQAKAFWNPDKTQFWTDSG---VDLVIDRREGYF
:: :.
Mb1683|M.bovis_AF2122/97 VTDVDGRTYIDLLGGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEP
Rv1655|M.tuberculosis_H37Rv VTDVDGRTYIDLLGGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEP
MLBr_01409|M.leprae_Br4923 VTDVDSNTYLDLLGGIAVNVLGHRHPAVIEAVTHQITTLGHTSNLYATEP
MAV_3115|M.avium_104 VTDVDGNTYLDLLGGIAVNVLGHRHPAIIEAVTKQLSTLGHTSNLYATEP
MMAR_2465|M.marinum_M VTDVDGKNYVDLLGGIAVNILGHRHPAVIEAVTQQMSTLGHTSNLYATEP
MUL_1647|M.ulcerans_Agy99 VTDVDGKNYVDLLGGIAVNILGHRHPAVIEAVTQQMSTLGHTSNLYATEP
TH_3529|M.thermoresistible__bu VTDADGKSYLDLLGGIAVNILGHRHPAVIEAVTRQLNTLGHTSNLYATEP
MAB_2279|M.abscessus_ATCC_1997 LTTAEGTRFLN-AGGYGVALAGYRHPVVVDHIRRQLDQHPVASRMFYEPA
Mflv_1243|M.gilvum_PYR-GCK LWDMTGRRLIDMHLNGGTYNLGHRNPEVMQAITDGMAHFDVGNHHFPSVA
Mvan_5565|M.vanbaalenii_PYR-1 LWDMSGRRLIDMHLNGGTYNLGHRNPEVMQAITDGMQYFDVGNHHFPSVA
MSMEG_0277|M.smegmatis_MC2_155 LWDMSGRRLIDLHLNGGTYNIGHRNPEVMQAISEGMAHFDVGNHHFPSVA
: . :: . .. *:*:* ::: : : .. : .
Mb1683|M.bovis_AF2122/97 GIALAEELVALLGADQR--------------TRVFFCNSGAEANEAAFKL
Rv1655|M.tuberculosis_H37Rv GIALAEELVALLGADQR--------------TRVFFCNSGAEANEAAFKL
MLBr_01409|M.leprae_Br4923 SITLAEELVALLGADTQ--------------TRVFFCNSGTEANELAFKL
MAV_3115|M.avium_104 SLALAEELVALLGAGHEPAGHESGGTESAGTARVFFCNSGTEANEVAFKL
MMAR_2465|M.marinum_M TIALAEELVTLLGAEAP--------------ARVFFCNSGTEANEAAFKL
MUL_1647|M.ulcerans_Agy99 TNALAEELVTLLGAEAP--------------ARVFFCNSGTEANEAAFKL
TH_3529|M.thermoresistible__bu GVALAEALVGHLGAPAR----------------VFFCNSGAEANEVAFKI
MAB_2279|M.abscessus_ATCC_1997 AAEAAAALAAVTPKGLD---------------RIHFACSGAEATEAAMKL
Mflv_1243|M.gilvum_PYR-GCK RTALAQKLIESAPQSLS---------------KVAFGSGGGEAIDIALKS
Mvan_5565|M.vanbaalenii_PYR-1 RTALAQKLIESAPASLS---------------KVAFGSGGGEAIDIALKS
MSMEG_0277|M.smegmatis_MC2_155 RTALAKRLVETAPASIK---------------KVAFGSGGGEAIDIALKS
* * : * .* ** : *:*
Mb1683|M.bovis_AF2122/97 SRLTG-RTKLVAAHDAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYG
Rv1655|M.tuberculosis_H37Rv SRLTG-RTKLVAAHDAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYG
MLBr_01409|M.leprae_Br4923 SRLTG-RTKLVAAQAAFHGRTMGSLALTGQPAKQAAFEPLPGHVTHVPYG
MAV_3115|M.avium_104 SRLTG-RTKLVAAQEAFHGRTMGSLALTGQPSKQAPFAPLPGDVTHVPYG
MMAR_2465|M.marinum_M SRLTG-RTKLVAAQEGFHGRTMGSLALTGQPAKQAPFEPLPGYVTHVPYG
MUL_1647|M.ulcerans_Agy99 SRLTG-RTKLVAAQEGFHGRTMGSLALTGQPAKQAPFEPLPGYVTHVPYG
TH_3529|M.thermoresistible__bu SRLTG-RTKVVAAEGGFHGRTMGALALTGQPSKREPFEPLPGDVIHVPYG
MAB_2279|M.abscessus_ATCC_1997 ARLNG-CRRFISMTGGYHGKTLGALSLTAREVFQAPFRPLLPGVSHVPYG
Mflv_1243|M.gilvum_PYR-GCK ARHATQRRKIVSIVKAYHGHTGLAVATGDDRFAKLFLADRPDEFVQVPFG
Mvan_5565|M.vanbaalenii_PYR-1 ARHATQRRKIVSIVKAYHGHTGLAVATGDDRFAKLFLADRPDEFVQVPFG
MSMEG_0277|M.smegmatis_MC2_155 ARHGTQRRKIVSVIKAYHGHTGLAVATGDDRFAKLFLADRPDEFIQVPFG
:* :.:: .:**:* ::: : : . :* :*
Mb1683|M.bovis_AF2122/97 DVDAL--AAAVDDHTAAVFLEPIMGESGVVVPPAGYLAAARDITARRGAL
Rv1655|M.tuberculosis_H37Rv DVDAL--AAAVDDHTAAVFLEPIMGESGVVVPPAGYLAAARDITARRGAL
MLBr_01409|M.leprae_Br4923 QVDAL--AAAVDNDTAAVFLEPIMGESGVIVPPEGYLAAARDITTRHGAL
MAV_3115|M.avium_104 DTEAL--AAAVDDGTAAVFLEPIMGESGVVVPPEGYLAAARDITARHGAL
MMAR_2465|M.marinum_M DIDAL--AAAVDDETAAVFLEPIMGESGVVVPPEGYLLAAREITARHGAL
MUL_1647|M.ulcerans_Agy99 DIDAL--AAAVDDETAAVFLEPIMGESGVVVPPEGYLLAAREITARHGAL
TH_3529|M.thermoresistible__bu DAGAL--ERAVDDTTAAVFLEPIMGEGGVVVPPAGYLAAAREVTARHGAL
MAB_2279|M.abscessus_ATCC_1997 DLAALRTELAMSTARACVIVEPVQGEAGVIIPPDGYLAGLRAVCDEYGAL
Mflv_1243|M.gilvum_PYR-GCK DTDAM-EQALRGRDVAAVIMETIPATYGFPLPPIGYLEAVKDLCVRYDAL
Mvan_5565|M.vanbaalenii_PYR-1 DTDAM-EQALRGRDVAAVIMETIPATYGFPLPPIGYLEAVKDLCVRYDAL
MSMEG_0277|M.smegmatis_MC2_155 DIGAM-EAALAGGDVAAVIMETIPATYGFPLPPPGYLEAVKALTERHGTL
: *: . *.*::*.: . *. :** *** . : : . .:*
Mb1683|M.bovis_AF2122/97 LVLDEVQTGMGRTGAFFAHQHDGITPDVVTLAKGLGGG-LPIGACLAVGP
Rv1655|M.tuberculosis_H37Rv LVLDEVQTGMGRTGAFFAHQHDGITPDVVTLAKGLGGG-LPIGACLAVGP
MLBr_01409|M.leprae_Br4923 LVIDEVQTGIGRTGAFFAHQHDSITPDVVTLAKGLGGG-LPIGAFLATGP
MAV_3115|M.avium_104 LVLDEVQTGMGRTGTFFAHQHDGITPDVVTLAKGLGGG-LPIGACLAVGP
MMAR_2465|M.marinum_M LVLDEVQTGIGRTGAFYAHQHDGITPDVVTLAKGLGGG-LPIGACLAIGP
MUL_1647|M.ulcerans_Agy99 LVLDEVQTGIGRTGAFYAHQHDGITPDVVTLAKGLGGG-LPIGACLAIGP
TH_3529|M.thermoresistible__bu LVLDEVQTGIGRTGTFFAHQHDGITPDVVTLAKGLGGG-LPIGACLAVGP
MAB_2279|M.abscessus_ATCC_1997 LVADEIQTGLGRLGQWWGVDRERVIPDILLSGKMLGGGVMPVSAAITTTE
Mflv_1243|M.gilvum_PYR-GCK YIADEVQTGLMRTGEMWGITKHGIEPDIMVTGKGLSGGMYPITAALLSDR
Mvan_5565|M.vanbaalenii_PYR-1 YIADEVQTGLMRTGEMWCISKHGIEPDIMVTGKGLSGGMYPITAALLSDR
MSMEG_0277|M.smegmatis_MC2_155 YIADEVQTGLMRTGELWGITKHGIDPDIIVTGKGLSGGMYPISAVLLGER
: **:***: * * : :. : **:: .* *.** *: * :
Mb1683|M.bovis_AF2122/97 AAELLTPG--LHGSTFGGNPVCAAAALAVLRVLASDGLVRRAEVLGKSLR
Rv1655|M.tuberculosis_H37Rv AAELLTPG--LHGSTFGGNPVCAAAALAVLRVLASDGLVRRAEVLGKSLR
MLBr_01409|M.leprae_Br4923 AAELLTLG--LHGSTFGGNPVCTAAALAVLRVLATQGLVRRAEVLGDSMR
MAV_3115|M.avium_104 AADLLTPG--LHGSTFGGNPVCAAAALAVLRVLASENLVRHAEVLGKSLH
MMAR_2465|M.marinum_M AAELLTPG--LHGSTFGGNPVCAAAALAVLRVVASEGLVRRAETLGKTFK
MUL_1647|M.ulcerans_Agy99 AAELLTPG--LHGSTFGGNPVCAAAALAVLRVVASEGLVCRAETLGKTFK
TH_3529|M.thermoresistible__bu AGDLLTPG--MHGSTFGGNPVCTAAAVAVLGALAEGDLINRADVLGKKLS
MAB_2279|M.abscessus_ATCC_1997 CFRQLDRDPYLHTSTFSTSPLGMAAVSGALRVIAEERLVERAAELGQILL
Mflv_1243|M.gilvum_PYR-GCK AAQWLHQDGFGHISTFGGAELGCVAALKTLEITSRPEVRSSVHYIADIFA
Mvan_5565|M.vanbaalenii_PYR-1 AAQWLDQDGFGHISTFGGAELGCVAALKTLEISSRPEVRSSVHYIADIFA
MSMEG_0277|M.smegmatis_MC2_155 AARWLDQDGFGHISTFGGAELGCVAAIKTLEICTRPEVRSMVHYIADIFD
. * . * ***. : .*. .* : : . :.. :
Mb1683|M.bovis_AF2122/97 HGIEALGHPLID----HVRGRGLLLGIALTAPHAKDAEATARDAGYLVN-
Rv1655|M.tuberculosis_H37Rv HGIEALGHPLID----HVRGRGLLLGIALTAPHAKDAEATARDAGYLVN-
MLBr_01409|M.leprae_Br4923 IGIESLSHPLID----QVRGRGLLLGIVLTAPRAKDIEKAARDAGFLVN-
MAV_3115|M.avium_104 QGIEGLGHPLVD----HVRGRGLLCGVVLTAPRAKDAEAAAREAGFLVN-
MMAR_2465|M.marinum_M QGVESLSHPLID----HVRGRGLLLGIVLAGPHAKAAEVAAREAGYLVN-
MUL_1647|M.ulcerans_Agy99 QGVESLSHPLID----HVRGRGLLLGIVLAGPHAKAAEVAAREVGYLVN-
TH_3529|M.thermoresistible__bu HGIEELRHPLVD----HVRGRGLLCGVVLNAPQAKAAEAAARDAGFLVN-
MAB_2279|M.abscessus_ATCC_1997 DRISSASAQWFGPDQVQVRGQGLLIGIEFTDASIVGDLLIELISNNIIVN
Mflv_1243|M.gilvum_PYR-GCK QGLAAIQADHPG-WFVGIRQNGVVIGLEFDHPEGAKFVMRELYQNGVWAI
Mvan_5565|M.vanbaalenii_PYR-1 KGLSRIQADYPG-WFVGIRQNGVVIGLEFDHPEGAKFVMRELYQNGVWAI
MSMEG_0277|M.smegmatis_MC2_155 TGLRRIQADYPD-WFVGIRQNGVVIGLEFDHPEGAKFVMRELYENGVWAI
: . :* .*:: *: : . . :
Mb1683|M.bovis_AF2122/97 --AAAPDVIRLAPPLIIAEAQLDGFVAALPAILDRAVGAP------
Rv1655|M.tuberculosis_H37Rv --AAAPDVIRLAPPLIIAEAQLDGFVAALPAILDRAVGAP------
MLBr_01409|M.leprae_Br4923 --ATAPEVIRLAPPLIITESQIDSFITALPGILDASIAELGKKA--
MAV_3115|M.avium_104 --AAAPGVIRLAPPLIVTEAQIDGFIAALPGILDRAAA--------
MMAR_2465|M.marinum_M --GAAPNVVRLAPPLVITEPQLEGFVAALPGILDTAGASA------
MUL_1647|M.ulcerans_Agy99 --GAAPNVVRLAPPLVITEPQLEGFVAALPGILDNAGASA------
TH_3529|M.thermoresistible__bu --AAAPEVIRLAPPLIITETQIDEFLAALPGVLDTAKEAS------
MAB_2279|M.abscessus_ATCC_1997 HSLNSDHVLRLTPPAVITDDDVEFLVQGYLNAIEQVAIRAKAALPG
Mflv_1243|M.gilvum_PYR-GCK FSTLDPRVLQFKPGLLLNRDLCEDVLDRVEVAVGRAKLAATGRRKP
Mvan_5565|M.vanbaalenii_PYR-1 FSTLDPRVLQFKPGLLLNRDLCEDVLDRVEVAVGRAKLAATGRRKP
MSMEG_0277|M.smegmatis_MC2_155 FSTLDPRVLQFKPGLLLSRELCEDVLDRLEVAVGRARATVTGRRNG
*::: * :: : .: :