For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGASTTTATMRQRWQAVMMNNYGTPPIALASGDGAVVTDVDGRTYIDLLGGIAVNVLGHRHPAVIEAVT RQMSTLGHTSNLYATEPGIALAEELVALLGADQRTRVFFCNSGAEANEAAFKLSRLTGRTKLVAAHDAFH GRTMGSLALTGQPAKQTPFAPLPGDVTHVGYGDVDALAAAVDDHTAAVFLEPIMGESGVVVPPAGYLAAA RDITARRGALLVLDEVQTGMGRTGAFFAHQHDGITPDVVTLAKGLGGGLPIGACLAVGPAAELLTPGLHG STFGGNPVCAAAALAVLRVLASDGLVRRAEVLGKSLRHGIEALGHPLIDHVRGRGLLLGIALTAPHAKDA EATARDAGYLVNAAAPDVIRLAPPLIIAEAQLDGFVAALPAILDRAVGAP
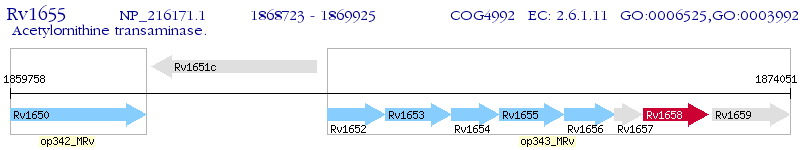
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1655 | argD | - | 100% (400) | acetylornithine aminotransferase |
| M. tuberculosis H37Rv | Rv3329 | - | 9e-51 | 34.94% (415) | hypothetical protein Rv3329 |
| M. tuberculosis H37Rv | Rv2589 | gabT | 1e-50 | 35.05% (428) | 4-aminobutyrate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1683 | argD | 0.0 | 100.00% (400) | acetylornithine aminotransferase |
| M. gilvum PYR-GCK | Mflv_3527 | argD | 1e-180 | 81.46% (383) | acetylornithine aminotransferase |
| M. leprae Br4923 | MLBr_01409 | argD | 0.0 | 82.87% (397) | acetylornithine aminotransferase |
| M. abscessus ATCC 19977 | MAB_2339 | argD | 1e-164 | 73.74% (396) | acetylornithine aminotransferase |
| M. marinum M | MMAR_2465 | argD | 0.0 | 84.14% (391) | acetylornithine aminotransferase ArgD |
| M. avium 104 | MAV_3115 | argD | 0.0 | 83.21% (405) | acetylornithine aminotransferase |
| M. smegmatis MC2 155 | MSMEG_3773 | argD | 1e-174 | 77.26% (387) | acetylornithine aminotransferase |
| M. thermoresistible (build 8) | TH_3529 | - | 1e-178 | 79.33% (387) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_1647 | argD | 0.0 | 83.38% (391) | acetylornithine aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_3311 | argD | 1e-179 | 79.37% (383) | acetylornithine aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3527|M.gilvum_PYR-GCK --MTGEHRPGLLDRWSAVMMNNYGTPPLALASGQGAVVTDADGKSYIDLL
Mvan_3311|M.vanbaalenii_PYR-1 --MT-----ELLNRWSAVMMNNYGTPPLALAGGDGAVVTDVDGKAYLDLL
MSMEG_3773|M.smegmatis_MC2_155 --------MTLQSRWEAVMMNNYGTPPLSLVSGEGAVVTDADGREYLDLL
TH_3529|M.thermoresistible__bu -VSGSKGRHTLLDRWQGVMMNNYGTPPLALVSGDGAVVTDADGKSYLDLL
Rv1655|M.tuberculosis_H37Rv MTGASTTTATMRQRWQAVMMNNYGTPPIALASGDGAVVTDVDGRTYIDLL
Mb1683|M.bovis_AF2122/97 MTGASTTTATMRQRWQAVMMNNYGTPPIALASGDGAVVTDVDGRTYIDLL
MLBr_01409|M.leprae_Br4923 MTPTQTNTATMQQRWETVMMNNYGTPPIVLASGNGAVVTDVDSNTYLDLL
MAV_3115|M.avium_104 ---MVTNTDALRRRWEAVMMHNYGTPPVALASGEGAVVTDVDGNTYLDLL
MMAR_2465|M.marinum_M ----MTHTEDTRARWEAVMMNNYGTPPVALASGEGAVVTDVDGKNYVDLL
MUL_1647|M.ulcerans_Agy99 ----MTHTEDTRARWEAVMMNNYGTPPVALASGEGAVVTDVDGKNYVDLL
MAB_2339|M.abscessus_ATCC_1997 MTEAPDSTSRAQQRWEHVMMNNYGTPPIVLAEGEGAVVTDVDGKRYLDLL
**. ***:******: *. *:******.*.. *:***
Mflv_3527|M.gilvum_PYR-GCK GGIAVNVLGHRHPAVIEAVTRQMNTLGHTSNLYATEPGVALAEALVDLLG
Mvan_3311|M.vanbaalenii_PYR-1 GGIAVNVLGHRHPAVIEAVTRQMNTLGHTSNLYATEPGVALAESLVGLLG
MSMEG_3773|M.smegmatis_MC2_155 GGIAVNLLGHRHPAVIEAVTTQLDTLGHTSNLYATEPGIALAEALVGQLG
TH_3529|M.thermoresistible__bu GGIAVNILGHRHPAVIEAVTRQLNTLGHTSNLYATEPGVALAEALVGHLG
Rv1655|M.tuberculosis_H37Rv GGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEPGIALAEELVALLG
Mb1683|M.bovis_AF2122/97 GGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEPGIALAEELVALLG
MLBr_01409|M.leprae_Br4923 GGIAVNVLGHRHPAVIEAVTHQITTLGHTSNLYATEPSITLAEELVALLG
MAV_3115|M.avium_104 GGIAVNVLGHRHPAIIEAVTKQLSTLGHTSNLYATEPSLALAEELVALLG
MMAR_2465|M.marinum_M GGIAVNILGHRHPAVIEAVTQQMSTLGHTSNLYATEPTIALAEELVTLLG
MUL_1647|M.ulcerans_Agy99 GGIAVNILGHRHPAVIEAVTQQMSTLGHTSNLYATEPTNALAEELVTLLG
MAB_2339|M.abscessus_ATCC_1997 GGIAVNVLGHRNQSVIDAVTQQLSTLGHTSNLYATGPAIELSEALVARLG
******:****: ::*:*** *: *********** * *:* ** **
Mflv_3527|M.gilvum_PYR-GCK VPAK----------------VFFCNSGTEANEVAFKITRLTGRTKLVAAQ
Mvan_3311|M.vanbaalenii_PYR-1 APAK----------------AFFCNSGTEANEAAFKITRLTGRTKVVAAE
MSMEG_3773|M.smegmatis_MC2_155 TQAR----------------VFFCNSGTEANEVAFKITRLTGKTKIVAAE
TH_3529|M.thermoresistible__bu APAR----------------VFFCNSGAEANEVAFKISRLTGRTKVVAAE
Rv1655|M.tuberculosis_H37Rv ADQR--------------TRVFFCNSGAEANEAAFKLSRLTGRTKLVAAH
Mb1683|M.bovis_AF2122/97 ADQR--------------TRVFFCNSGAEANEAAFKLSRLTGRTKLVAAH
MLBr_01409|M.leprae_Br4923 ADTQ--------------TRVFFCNSGTEANELAFKLSRLTGRTKLVAAQ
MAV_3115|M.avium_104 AGHEPAGHESGGTESAGTARVFFCNSGTEANEVAFKLSRLTGRTKLVAAQ
MMAR_2465|M.marinum_M AEAP--------------ARVFFCNSGTEANEAAFKLSRLTGRTKLVAAQ
MUL_1647|M.ulcerans_Agy99 AEAP--------------ARVFFCNSGTEANEAAFKLSRLTGRTKLVAAQ
MAB_2339|M.abscessus_ATCC_1997 VPGR----------------VFLCNSGTEANEAAFKLTRLTGRTKVVAAE
. .*:****:**** ***::****:**:***.
Mflv_3527|M.gilvum_PYR-GCK GAFHGRTMGSLALTGQPAKQAPFEPLPGDVTHVPYGDVDALRAVVDSETA
Mvan_3311|M.vanbaalenii_PYR-1 GAFHGRTMGSLALTGQPSKRAPFEPLPGGVTHVPYGDIDELRRVVDDQTA
MSMEG_3773|M.smegmatis_MC2_155 GAFHGRTMGSLALTGQPSKQAPFEPLPGNVMHVPYGDVAALEAAVDDQTA
TH_3529|M.thermoresistible__bu GGFHGRTMGALALTGQPSKREPFEPLPGDVIHVPYGDAGALERAVDDTTA
Rv1655|M.tuberculosis_H37Rv DAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYGDVDALAAAVDDHTA
Mb1683|M.bovis_AF2122/97 DAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYGDVDALAAAVDDHTA
MLBr_01409|M.leprae_Br4923 AAFHGRTMGSLALTGQPAKQAAFEPLPGHVTHVPYGQVDALAAAVDNDTA
MAV_3115|M.avium_104 EAFHGRTMGSLALTGQPSKQAPFAPLPGDVTHVPYGDTEALAAAVDDGTA
MMAR_2465|M.marinum_M EGFHGRTMGSLALTGQPAKQAPFEPLPGYVTHVPYGDIDALAAAVDDETA
MUL_1647|M.ulcerans_Agy99 EGFHGRTMGSLALTGQPAKQAPFEPLPGYVTHVPYGDIDALAAAVDDETA
MAB_2339|M.abscessus_ATCC_1997 GAFHGRTMGSLALTGQPAKQAPFEPLPAGVLHVPYGDLAAIEAAVDGDTA
.*******:*******:*: .* ***. * ** **: : .**. **
Mflv_3527|M.gilvum_PYR-GCK AVFLEPIMGEGGVVVPPAGYLVAAREITAEHGALLILDEVQTGVGRTGAF
Mvan_3311|M.vanbaalenii_PYR-1 AVFLEPIMGEGGVVVPPAGYLVAAREITAAHGALLVLDEVQTGVGRTGAF
MSMEG_3773|M.smegmatis_MC2_155 AVFLEPIMGEGGVVVPPAGYLVAAREITSKHGALLVLDEVQTGVGRTGAF
TH_3529|M.thermoresistible__bu AVFLEPIMGEGGVVVPPAGYLAAAREVTARHGALLVLDEVQTGIGRTGTF
Rv1655|M.tuberculosis_H37Rv AVFLEPIMGESGVVVPPAGYLAAARDITARRGALLVLDEVQTGMGRTGAF
Mb1683|M.bovis_AF2122/97 AVFLEPIMGESGVVVPPAGYLAAARDITARRGALLVLDEVQTGMGRTGAF
MLBr_01409|M.leprae_Br4923 AVFLEPIMGESGVIVPPEGYLAAARDITTRHGALLVIDEVQTGIGRTGAF
MAV_3115|M.avium_104 AVFLEPIMGESGVVVPPEGYLAAARDITARHGALLVLDEVQTGMGRTGTF
MMAR_2465|M.marinum_M AVFLEPIMGESGVVVPPEGYLLAAREITARHGALLVLDEVQTGIGRTGAF
MUL_1647|M.ulcerans_Agy99 AVFLEPIMGESGVVVPPEGYLLAAREITARHGALLVLDEVQTGIGRTGAF
MAB_2339|M.abscessus_ATCC_1997 AVFLEPIMGEGGVVVPPEGYLAGVREITARHGALLVLDEVQTGIGRTGTF
**********.**:*** *** ..*::*: :****::******:****:*
Mflv_3527|M.gilvum_PYR-GCK FAHRHDGITPDVVTLAKGLGGGLPIGACLAIGDTAELLTPGLHGSTFGGN
Mvan_3311|M.vanbaalenii_PYR-1 YAHQHDGITPDIVTLAKGLGGGLPIGACLAIGDTADLLTPGLHGSTFGGN
MSMEG_3773|M.smegmatis_MC2_155 FAHQHDGIVPDVVTMAKGLGGGLPIGACLAVGATGDLLTPGLHGSTFGGN
TH_3529|M.thermoresistible__bu FAHQHDGITPDVVTLAKGLGGGLPIGACLAVGPAGDLLTPGMHGSTFGGN
Rv1655|M.tuberculosis_H37Rv FAHQHDGITPDVVTLAKGLGGGLPIGACLAVGPAAELLTPGLHGSTFGGN
Mb1683|M.bovis_AF2122/97 FAHQHDGITPDVVTLAKGLGGGLPIGACLAVGPAAELLTPGLHGSTFGGN
MLBr_01409|M.leprae_Br4923 FAHQHDSITPDVVTLAKGLGGGLPIGAFLATGPAAELLTLGLHGSTFGGN
MAV_3115|M.avium_104 FAHQHDGITPDVVTLAKGLGGGLPIGACLAVGPAADLLTPGLHGSTFGGN
MMAR_2465|M.marinum_M YAHQHDGITPDVVTLAKGLGGGLPIGACLAIGPAAELLTPGLHGSTFGGN
MUL_1647|M.ulcerans_Agy99 YAHQHDGITPDVVTLAKGLGGGLPIGACLAIGPAAELLTPGLHGSTFGGN
MAB_2339|M.abscessus_ATCC_1997 FAHQQAGITPDVVTLAKGLGGGLPIGACIATGAAADLLTPGMHGSTFGGN
:**:: .*.**:**:************ :* * :.:*** *:********
Mflv_3527|M.gilvum_PYR-GCK PVCTAAALAVLKVLADDGLVERADILGKTIAHGIEALNHPLVDHVRGLGL
Mvan_3311|M.vanbaalenii_PYR-1 PVCTAAALAVLKVLADEGLVERADVLGKTIAHGVESLGHPLVDHVRGRGL
MSMEG_3773|M.smegmatis_MC2_155 PVCTAAGLAVLKTLAAEDLVARAGVLGKTLSHGIEELGHPLVDKVRGKGL
TH_3529|M.thermoresistible__bu PVCTAAAVAVLGALAEGDLINRADVLGKKLSHGIEELRHPLVDHVRGRGL
Rv1655|M.tuberculosis_H37Rv PVCAAAALAVLRVLASDGLVRRAEVLGKSLRHGIEALGHPLIDHVRGRGL
Mb1683|M.bovis_AF2122/97 PVCAAAALAVLRVLASDGLVRRAEVLGKSLRHGIEALGHPLIDHVRGRGL
MLBr_01409|M.leprae_Br4923 PVCTAAALAVLRVLATQGLVRRAEVLGDSMRIGIESLSHPLIDQVRGRGL
MAV_3115|M.avium_104 PVCAAAALAVLRVLASENLVRHAEVLGKSLHQGIEGLGHPLVDHVRGRGL
MMAR_2465|M.marinum_M PVCAAAALAVLRVVASEGLVRRAETLGKTFKQGVESLSHPLIDHVRGRGL
MUL_1647|M.ulcerans_Agy99 PVCAAAALAVLRVVASEGLVCRAETLGKTFKQGVESLSHPLIDHVRGRGL
MAB_2339|M.abscessus_ATCC_1997 PVCVAAALAVLRVLDEQDLVDHAAAAGKSLSHSVEELNHPLVDRVRGAGL
***.**.:*** .: .*: :* *..: .:* * ***:*:*** **
Mflv_3527|M.gilvum_PYR-GCK LRGIVLTADAAKSVEAEARDAGFLVNAAAADVIRLAPPLVITDAEVESFL
Mvan_3311|M.vanbaalenii_PYR-1 LRGIVLTADAAKPVETAAREAGFLVNAAAPAIVRLAPPLVITEAQVETFL
MSMEG_3773|M.smegmatis_MC2_155 LQGIVLTVPSAKAVETAARDAGFLVNAAAPEVVRLAPPLIITEGQIEAFI
TH_3529|M.thermoresistible__bu LCGVVLNAPQAKAAEAAARDAGFLVNAAAPEVIRLAPPLIITETQIDEFL
Rv1655|M.tuberculosis_H37Rv LLGIALTAPHAKDAEATARDAGYLVNAAAPDVIRLAPPLIIAEAQLDGFV
Mb1683|M.bovis_AF2122/97 LLGIALTAPHAKDAEATARDAGYLVNAAAPDVIRLAPPLIIAEAQLDGFV
MLBr_01409|M.leprae_Br4923 LLGIVLTAPRAKDIEKAARDAGFLVNATAPEVIRLAPPLIITESQIDSFI
MAV_3115|M.avium_104 LCGVVLTAPRAKDAEAAAREAGFLVNAAAPGVIRLAPPLIVTEAQIDGFI
MMAR_2465|M.marinum_M LLGIVLAGPHAKAAEVAAREAGYLVNGAAPNVVRLAPPLVITEPQLEGFV
MUL_1647|M.ulcerans_Agy99 LLGIVLAGPHAKAAEVAAREVGYLVNGAAPNVVRLAPPLVITEPQLEGFV
MAB_2339|M.abscessus_ATCC_1997 LLGIVLTEQRAKAAETAAREAGFLVNAAAPDVLRLAPPLIITDEQIGEFV
* *:.* ** * **:.*:***.:*. ::******:::: :: *:
Mflv_3527|M.gilvum_PYR-GCK AALPRILDKAAQS-----
Mvan_3311|M.vanbaalenii_PYR-1 SALPAILDKAAQS-----
MSMEG_3773|M.smegmatis_MC2_155 TALPAVLDTAAEDS----
TH_3529|M.thermoresistible__bu AALPGVLDTAKEAS----
Rv1655|M.tuberculosis_H37Rv AALPAILDRAVGAP----
Mb1683|M.bovis_AF2122/97 AALPAILDRAVGAP----
MLBr_01409|M.leprae_Br4923 TALPGILDASIAELGKKA
MAV_3115|M.avium_104 AALPGILDRAAA------
MMAR_2465|M.marinum_M AALPGILDTAGASA----
MUL_1647|M.ulcerans_Agy99 AALPGILDNAGASA----
MAB_2339|M.abscessus_ATCC_1997 TALPGILDTAAEQEQA--
:*** :** :