For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLLTRFIKMQLVIFLTLTLVALVVLALFYLRLPTWAGLGMYKLNADLPNSGGLYATANVTYRGTTIGKVT SVEPSESGARVEMNIYDRYKIPADATANVHSVSAVGEQFIDLTSDSGGGAYFQPGDTITKATVPAEVGPA LDAAEKGLAVLPKEKIGTLLDEAATAFGGLGPSLQRLVDSTQAIAGDFRANIDPVNDIIENSGPIIDSQV NSGDAIQRWAANLNTLAAQSAQNDEALRSGLQQAAPTADQLNAVFSDVRESLPQTLANLEIVIDMLKRYN KNVEQVLVALPQGAAVAQTGTIFAPEGLLHFGLGINAPPPCLTGFLPASQWRSPADTRTEPLPSGLYCKI PKDAPNAVRGARNYPCADVPGKRAATPRECRSDEPYQPLGTNPWYGDPDQIRNCPAPGARCDQPVDPGRV IPAPSINNGLNPLPASQLPPPEVSSGPSSDPLTAPRGGTVTCSGQQPNPCIYTPAAGATATYNPASGEVV GPGGVKYSVTNSNTPGDDGWKEMLAPAS
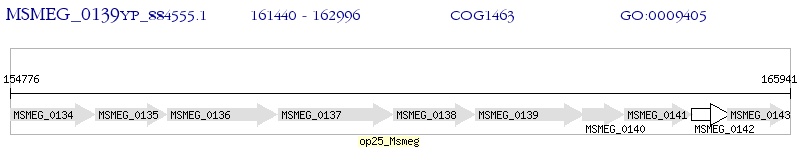
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0139 | - | - | 100% (518) | mce-family protein mce1f |
| M. smegmatis MC2 155 | MSMEG_2860 | - | 4e-90 | 37.98% (524) | virulence factor mce family protein |
| M. smegmatis MC2 155 | MSMEG_4785 | - | 8e-88 | 35.61% (570) | mce-family protein mce1f |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0180 | mce1F | 0.0 | 69.23% (520) | MCE-family protein MCE1F |
| M. gilvum PYR-GCK | Mflv_0706 | - | 0.0 | 70.58% (520) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0174 | mce1F | 0.0 | 69.04% (520) | MCE-family protein MCE1F |
| M. leprae Br4923 | MLBr_02594 | mce1F | 0.0 | 64.23% (520) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4148c | - | 4e-84 | 37.70% (496) | MCE-family protein |
| M. marinum M | MMAR_0417 | mce1F | 0.0 | 66.41% (518) | MCE-family protein Mce1F |
| M. avium 104 | MAV_4546 | - | 0.0 | 66.86% (516) | mce-family protein mce2f |
| M. thermoresistible (build 8) | TH_2681 | - | 0.0 | 73.03% (519) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1067 | mce1F | 0.0 | 66.02% (518) | MCE-family protein Mce1F |
| M. vanbaalenii PYR-1 | Mvan_0139 | - | 0.0 | 71.35% (520) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0180|M.bovis_AF2122/97 -MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVGIGQYTLKADLPAS
Rv0174|M.tuberculosis_H37Rv -MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVGIGQYTLKADLPAS
MMAR_0417|M.marinum_M -MLTPFIKRQLIMFGTLTVLSLLVLGVYYLQIPALLGVGRYPLKAELPAS
MUL_1067|M.ulcerans_Agy99 -MLTPFIKRQLIMFGTLTVLSLLVLGVYYLQIPALLGVGRYPLKAELPAS
MLBr_02594|M.leprae_Br4923 -MLTPLIRRQLVLFAILTVISLTVLGVYYLQIPSLVGIGRYSLKAELSAT
MAV_4546|M.avium_104 -----MIKTQLVLLTAVAVAAVVVLGWYYLRIPSLAGIGRYTLYAELPQS
Mflv_0706|M.gilvum_PYR-GCK -MLTRFIKIQLIIFTILTVVALGVLGLYYLRLPSLVGVGQYTLYAELPRS
Mvan_0139|M.vanbaalenii_PYR-1 -MLTRFIKFQLITFTILTVVALGVLGLYYLRLPSLAGIGEYKLYAELERS
TH_2681|M.thermoresistible__bu -MLTRFIKMQLVIFTVLTVIALVALGWYYLRVPSLIGIGQYTLHAELPRS
MSMEG_0139|M.smegmatis_MC2_155 MLLTRFIKMQLVIFLTLTLVALVVLALFYLRLPTWAGLGMYKLNADLPNS
MAB_4148c|M.abscessus_ATCC_199 --MERLIKIQLAIFAAVTVISVALMAIFYLRVPTALGIGKHDVKAQFVAS
:*: ** : ::: :: :. :**::*: *:* : : *:: :
Mb0180|M.bovis_AF2122/97 GGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSIASNYKIPVDASANVH
Rv0174|M.tuberculosis_H37Rv GGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSIASNYKIPVDASANVH
MMAR_0417|M.marinum_M GGLYPTANVTYRGITIGKVTGVEPTATGVEATMSIDSRYKIPVDASANVH
MUL_1067|M.ulcerans_Agy99 GGLYPTANVTYRGITIGKVTGVEPTATGVEATMSIDSRYKIPVDASANVH
MLBr_02594|M.leprae_Br4923 GGLYPTANVTYRGIAIGKVTDVEPTATGAEATMSIDSRYKIPIDSAANVH
MAV_4546|M.avium_104 GGLYRTANVTYRGITIGKVTGVEPTERGARATMSIEDGYRIPADAAAHVH
Mflv_0706|M.gilvum_PYR-GCK GGLYPTGNVTYRGTQIGKVTEVEPTETGARATMSIDNEYKVPADATANVH
Mvan_0139|M.vanbaalenii_PYR-1 GGLYPTGNVTYRGTQIGKVTEVEPTETGARVTMSIGDEYKIPADVSANVH
TH_2681|M.thermoresistible__bu GGLYATANVTYRGQTIGKVTAVEPTPRGARATMSIGSEYKIPIDSTANVH
MSMEG_0139|M.smegmatis_MC2_155 GGLYATANVTYRGTTIGKVTSVEPSESGARVEMNIYDRYKIPADATANVH
MAB_4148c|M.abscessus_ATCC_199 GGLYQNANVTFRGVQIGRVTDVELTPEGVTATMRLNDNVKVPGNVVAAVK
**** ..***:** **:** ** : *. . * : . ::* : * *:
Mb0180|M.bovis_AF2122/97 SVSAVGEQYIDLVST-GAPGKYFSSGQTITKG--TVPSEIGPALDNSNRG
Rv0174|M.tuberculosis_H37Rv SVSAVGEQYIDLVST-GAPGKYFSSGQTITKG--TVPSEIGPALDNSNRG
MMAR_0417|M.marinum_M SVSAVGEQYLDLVST-GAPAKFLSPGQTITKG--TVPSEIGPALDTANRG
MUL_1067|M.ulcerans_Agy99 SVSAVGEQYLDLVST-GAPAKFLSPGQTITKG--TVPSEIGPALDTANRG
MLBr_02594|M.leprae_Br4923 SVSAVGEQYLDLVSS-GNPGKYLAPGQTITKS--KIPIEIGPALDSVNRG
MAV_4546|M.avium_104 SVSAVGEQYVDLVST-AGREPYLVDGQTIHKS--TVPSQIGPALDAANRG
Mflv_0706|M.gilvum_PYR-GCK SVSAIGEQYLDLVST-SGTTQYLSPGQTITEG--TVPSEVGPTIDAANDG
Mvan_0139|M.vanbaalenii_PYR-1 SVSAIGEQYLDLVSE-TGTDQYLSAGQTITKS--TVPAEVGPALDAANNG
TH_2681|M.thermoresistible__bu SVSAIGEQYLDLVPA-GTTDVYFSDGDTITES--TVPSEVGPALDAANEG
MSMEG_0139|M.smegmatis_MC2_155 SVSAVGEQFIDLTSD-SGGGAYFQPGDTITKA--TVPAEVGPALDAAEKG
MAB_4148c|M.abscessus_ATCC_199 SVSAIGEQYVDLIPPPTGAAGTLRSGDVIGADRTSIPTDVSTLLRQADGL
****:***::** . : *:.* .:* ::.. : :
Mb0180|M.bovis_AF2122/97 LAALPTEKIGLLLDETAQAVGGLGPALQRLVDSTQAIVGDFKTNIGDVND
Rv0174|M.tuberculosis_H37Rv LAALPTEKIGLLLDETAQAVGGLGPALQRLVDSTQAIVGDFKTNIGDVND
MMAR_0417|M.marinum_M LEVLPADKIPVLLDETAQAVGGLGPALQRLVDATQAIVGDFHTQINDVND
MUL_1067|M.ulcerans_Agy99 LEVLPADKIPVLLDETAQAVGGLGPALQRLVDATQAIVGDFHTQINDVND
MLBr_02594|M.leprae_Br4923 LAALPKEKIGSLLDETAQAVGGLGPALQRLVSSTQAIVDAFKTNITDVND
MAV_4546|M.avium_104 LAVLPRDKIASLLYETSQAVGGLGPSLRRLVDATQAIAHDFRGSIDHVDD
Mflv_0706|M.gilvum_PYR-GCK LAVLPKEKIDALLTETSEAVGGLGPSLQRLVDSTTNLAEGFRDNLPQVTD
Mvan_0139|M.vanbaalenii_PYR-1 LAVLPKEKIDGLLTETSAAVGGLGPALQRLVDSTTNVAEGFRDNLPQVND
TH_2681|M.thermoresistible__bu LQALPAEKIDSLLTETAAAVGGLGPSLQRLVDSTTAIARDFRDNLEPVGD
MSMEG_0139|M.smegmatis_MC2_155 LAVLPKEKIGTLLDEAATAFGGLGPSLQRLVDSTQAIAGDFRANIDPVND
MAB_4148c|M.abscessus_ATCC_199 VNTVADTRLREVLAEAFKAFNGSGQELSRLIDSSRLLIQEANTNYDSTTK
: .:. :: :* *: *..* * * **:.:: : . . . .
Mb0180|M.bovis_AF2122/97 IIENSGPILDSQVNTGDQIERWARKLNNLAAQTATRDQNVRSILSQAAPT
Rv0174|M.tuberculosis_H37Rv IIENSGPILDSQVNTGDQIERWARKLNNLAAQTATRDQNVRSILSQAAPT
MMAR_0417|M.marinum_M IIQNSGPILDSQVNSGDAISRWAHNLNVLATQTAERDQNVKSILTQAAPT
MUL_1067|M.ulcerans_Agy99 IIQNSGPILDSQVNSGDAISRWAHNLNVLATQTAERDQNVKSILTQAAPT
MLBr_02594|M.leprae_Br4923 IIQNSGPILDSQVKSDNAIERWVHNLNVLAAQSARQDQHLKSILSQAAPT
MAV_4546|M.avium_104 IVERSAPIIDSQADSADTLGRWAANLNTLAAQTARQDPALRSILANAAPT
Mflv_0706|M.gilvum_PYR-GCK IINNAAPILDSQVQSGDAIEQWSRNLNVIASQTAEQDAALRSGLQQAAPT
Mvan_0139|M.vanbaalenii_PYR-1 IITNSAPILQSQVTSGDAIEQWSRNLNVIASQTAEQDAALRSGLQQAAPT
TH_2681|M.thermoresistible__bu IIDNSAPILDSQVDSGDAIARWSRNVNILAAQAAQQDAALRSGLQQAAPT
MSMEG_0139|M.smegmatis_MC2_155 IIENSGPIIDSQVNSGDAIQRWAANLNTLAAQSAQNDEALRSGLQQAAPT
MAB_4148c|M.abscessus_ATCC_199 LIDQVGPLLDAQIRSGDNIRGSVSSLAALTEHFAKADPQLRQFLKVAPGA
:: . .*::::* : : : .: :: : * * ::. * *. :
Mb0180|M.bovis_AF2122/97 ADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHAGVEQLLVFLPQ--GAA
Rv0174|M.tuberculosis_H37Rv ADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHAGVEQLLVFLPQ--GAA
MMAR_0417|M.marinum_M ADQLNSVFSDVRDSLPQTLANLEVVFDLLKRYHKGLEQVLVFLPQ--GAS
MUL_1067|M.ulcerans_Agy99 ADQLNSVFSDVRDSLPQTLANLEVVFDLLKRYHKGLEQVLVFLPQ--GAS
MLBr_02594|M.leprae_Br4923 ADEVNTVFSDVRDSLPQTLANFEILIDMLKRYHHGVEQMLVFLPQ--GAS
MAV_4546|M.avium_104 AEQVRATFGGVRESLPQTLANLEVVIDMLKRYHNGVEQALVFLPQ--SGA
Mflv_0706|M.gilvum_PYR-GCK LDQVNTVFSDVQEALPQTLANLAVVNDMLKRYNKGIEQTLVIYPQ--GAS
Mvan_0139|M.vanbaalenii_PYR-1 LDQVTTVFSDVRDALPQTLANLAVINDMLKRYNKGIEQTLVIYPQ--GAS
TH_2681|M.thermoresistible__bu ADQLNAVFGGVRESLPQVLANLAVVSDMLKRYHKGLNQSLVILPQ--GAA
MSMEG_0139|M.smegmatis_MC2_155 ADQLNAVFSDVRESLPQTLANLEIVIDMLKRYNKNVEQVLVALPQ--GAA
MAB_4148c|M.abscessus_ATCC_199 SEQASQLFAGIRPSFPVLAASLANLGRVGVIYNKSIEQSLVILPALFSAL
:: *..:: ::* *.: : : *: .::* ** * ..
Mb0180|M.bovis_AF2122/97 IAQTVLTPTPGAAQLPL-APAINYPPPCLTGFLPASEWRSPADTSPRPLP
Rv0174|M.tuberculosis_H37Rv IAQTVLTPTPGAAQLPL-APAINYPPPCLTGFLPASEWRSPADTSPRPLP
MMAR_0417|M.marinum_M IAQTVAAPFPNMAALDM-ALAINQPPPCLTGFIPAEQWRSFADTSLQPLP
MUL_1067|M.ulcerans_Agy99 IAQTVAAPFPNMAALDM-ALAINQPPPCLTGFIPAEQWRSFADTSLQPLP
MLBr_02594|M.leprae_Br4923 IAQTASATFPNMAALDL-ALSINQPPPCLTGFIPAAEWRSPADTSLQPLP
MAV_4546|M.avium_104 IAQSVTAQSPGQAALGVGAISLNQPPPCLTGFLPASQWRAPADTSTAPLP
Mflv_0706|M.gilvum_PYR-GCK LLQMG-SIFQDGGLLHF-GLSINQPPPCMTGFLPASEWRSPADTSQAPLP
Mvan_0139|M.vanbaalenii_PYR-1 LLQMG-SIFEDGGLLHF-GLSINQPPPCMTGFLPASEWRSPADTSTAPLP
TH_2681|M.thermoresistible__bu IAQTTTAPFPGEALLHL-ALGINIPPPCLTGFLPPSQWRSPADTSPQPLP
MSMEG_0139|M.smegmatis_MC2_155 VAQTG-TIFAPEGLLHF-GLGINAPPPCLTGFLPASQWRSPADTRTEPLP
MAB_4148c|M.abscessus_ATCC_199 LTVAGGAPMDEGAKLDF-RLNLGDPPPCLTGFVPPPLMRTPADETLRELP
: : . * . :. ****:***:*. *: ** **
Mb0180|M.bovis_AF2122/97 SGTYCKIPQDAQLQ-VRGARNIPCVDVPGKRAATPKECRSKDPYVPLGTN
Rv0174|M.tuberculosis_H37Rv SGTYCKIPQDAQLQ-VRGARNIPCVDVLGKRAATPKECRSKDPYVPLGTN
MMAR_0417|M.marinum_M KGTYCKIPMDTPANSVRGSRNIPCVDVPGKRAATPRECRDPKPYVPAGTN
MUL_1067|M.ulcerans_Agy99 KGTYCKIPMDTPANSVRGSRNIPCVDVPGKRAATPRECRDPKPYVPAGTN
MLBr_02594|M.leprae_Br4923 VGTYCKIPMDTPANSVRGSRNIPCVDVPGKRAASPRECRDSKPYVPLGTN
MAV_4546|M.avium_104 AGTYCKIPMD-ATNVVRGARNYPCVDVPGKRAATPRECRSTEPYVPQGTN
Mflv_0706|M.gilvum_PYR-GCK SGMYCKIPKDYQGNSVRGARNYPCPDVPGKRAASPKECRSDEPYVPLGTN
Mvan_0139|M.vanbaalenii_PYR-1 SGMYCKIPKDYQGNAVRGARNYPCVDVPGKRAASPKECRSDEPYVPLGTN
TH_2681|M.thermoresistible__bu EGTYCKIPKDTQGNVVRGGRNYPCVDVPGKRAATPRECRSDEPYVPLGTN
MSMEG_0139|M.smegmatis_MC2_155 SGLYCKIPKDAP-NAVRGARNYPCADVPGKRAATPRECRSDEPYQPLGTN
MAB_4148c|M.abscessus_ATCC_199 RDMYCKTAQNDP-SAVRGARNYPCQEFPGKRAPTIQLCRDPRGYVPIGSN
. *** . : . ***.** ** :. ****.: : **. * * *:*
Mb0180|M.bovis_AF2122/97 PWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTGLNPAPADQVQ---
Rv0174|M.tuberculosis_H37Rv PWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTGLNPAPADQVQ---
MMAR_0417|M.marinum_M PWYGDPNQLLTCPAPGARCDQPVKPGLVAQAPSVNNGMNPAPADKLPPG-
MUL_1067|M.ulcerans_Agy99 PWYGDPNQLLTCPAPGARCDQSVKPGLVAQAPSVNNGMNPAPADKLPPG-
MLBr_02594|M.leprae_Br4923 PWFGDPNQLLTCPAPGARCDQPVKPGLVIPAPTVDNGLNPAPADRLP---
MAV_4546|M.avium_104 PWYGDPDQILTCPAPGARCDQPVKPGLVIPAPSVDNGLNPLPADRLP---
Mflv_0706|M.gilvum_PYR-GCK PWYGDPNQIVACPAPGARCDQRVAPGQVVPAPSINNGQNPLPADELPPPA
Mvan_0139|M.vanbaalenii_PYR-1 PWYGDPNQIVACPAPGARCDQRVAPGQVIPAPSVNNGQNPLPADQLPPPQ
TH_2681|M.thermoresistible__bu PWYGDPNQIRNCPAPGARCDQPVDPGKVIPAESVNNGMNPLPADRLPGP-
MSMEG_0139|M.smegmatis_MC2_155 PWYGDPDQIRNCPAPGARCDQPVDPGRVIPAPSINNGLNPLPASQLPPPE
MAB_4148c|M.abscessus_ATCC_199 PWRGPPAPYDTPVEDPRNILPPNKFPNIPPGAEPDPGTPTLPPPSPPAP-
** * * . : . : * . *.
Mb0180|M.bovis_AF2122/97 GTPPPVSDPLQRPG-SGTVQCNGQQPNPCVYTPTSGP---SAVYSPASGE
Rv0174|M.tuberculosis_H37Rv GTPPPVSDPLQRPG-SGTVQCNGQQPNPCVYTPTSGP---SAVYSPASGE
MMAR_0417|M.marinum_M GTPPPISDPLQRPG-SGTVQCNGQQPNPCVYTP-GPP---TAVYSPQSGE
MUL_1067|M.ulcerans_Agy99 GTPPPISDPLERPG-SGTVQCNGQQPNPCVYTP-GPP---TAVYSPQSGK
MLBr_02594|M.leprae_Br4923 GTPPPVSDPLSQPR-SGSVQCNGQQPNTCVYTPSGPP---SAVYSPQSGE
MAV_4546|M.avium_104 GTPPPISDPLQRPG-SGTVVCNGQQPNPCTYTP-------SALYDVRSGT
Mflv_0706|M.gilvum_PYR-GCK S-LAPVSDPVSPPG-SGTVRCSGQQPNPCTYTPAQGPPGTTAGYSPSSGE
Mvan_0139|M.vanbaalenii_PYR-1 S-LAPVSDPLSPPG-SGTVRCSGQQPNPCAYTPAQGPPGTTAGYSPSSGE
TH_2681|M.thermoresistible__bu --VPPVSDPLTPPGPSGMVECSGQQPNPCIYTPAPG----TAIYNPASGE
MSMEG_0139|M.smegmatis_MC2_155 VSSGPSSDPLTAPR-GGTVTCSGQQPNPCIYTPAAG---ATATYNPASGE
MAB_4148c|M.abscessus_ATCC_199 EVPPAGGDVMPVSAQSGPMAARAYDPKNGVFLDANNQ---PSVYAP----
. .* : . .* : . . :*: : .: *
Mb0180|M.bovis_AF2122/97 LVGPDGVKYAVANSSTTGDDGWKEMLAPAS-
Rv0174|M.tuberculosis_H37Rv LVGPDGVKYAVANSSTTGDDGWKEMLAPAS-
MMAR_0417|M.marinum_M LVGPDGVKYSVENSTKTGDDGWKEMLAPAG-
MUL_1067|M.ulcerans_Agy99 LVGPDGVKYSVENSTKTGDDGWKEMLAPAG-
MLBr_02594|M.leprae_Br4923 LVGPDGVAYAVENSTKTGDDGWKEMLAPAG-
MAV_4546|M.avium_104 VVGPDGVVYSVANSATIGDEGWKTMLGQAR-
Mflv_0706|M.gilvum_PYR-GCK VVGPDGVRYSVTNSTNPGDDGWKEMLAPAG-
Mvan_0139|M.vanbaalenii_PYR-1 VVGPDGVRYNVSNSNNPGDDGWKEMLAPAG-
TH_2681|M.thermoresistible__bu AVGPDGVKYSVSNSSNPGDDGWKEMLAPAG-
MSMEG_0139|M.smegmatis_MC2_155 VVGPGGVKYSVTNSNTPGDDGWKEMLAPAS-
MAB_4148c|M.abscessus_ATCC_199 ----------ALDRGTEAETWADLMLGPKPV
. : . .: . **.