For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSLRTDDDTWDIATSVGSTAVMVAAARAGETERDDALIRDPYAKILVAGAGTGVWETVLDSEFAAKIEN LDPEAAAIFAHMGNYQAVRTHFFDAYYREAADAGIRQIVILASGLDSRAYRLDWPAGTTVYEIDQPKVLE YKSATLREHGIEPVAQRREVPVDLRFDWPTALHDAGFDASLPTAWLAEGLLMYLPAEAQDRLFELVTELS APGSRIAVETAGVAATDRREAMRERFKKFADQLNLSSALDIQELVYDDPDRADVAEWLDAHGWRARGVHA LDEMRRLDRLVELPDDPDRAAFSTFVTAQRL
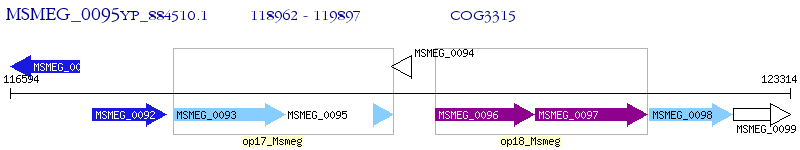
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0095 | - | - | 100% (311) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_0093 | - | 8e-80 | 55.48% (292) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1482 | - | 2e-74 | 49.51% (307) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0151 | - | 1e-111 | 66.13% (310) | hypothetical protein Mb0151 |
| M. gilvum PYR-GCK | Mflv_0743 | - | 1e-119 | 67.52% (311) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv0146 | - | 1e-111 | 66.13% (310) | hypothetical protein Rv0146 |
| M. leprae Br4923 | MLBr_02640 | - | 1e-108 | 63.87% (310) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_4606c | - | 8e-87 | 53.33% (300) | hypothetical protein MAB_4606c |
| M. marinum M | MMAR_0357 | - | 1e-114 | 65.50% (313) | O-methyltransferase |
| M. avium 104 | MAV_5149 | - | 1e-113 | 66.67% (312) | methyltransferase, putative, family protein |
| M. thermoresistible (build 8) | TH_4502 | - | 1e-128 | 72.41% (319) | PUTATIVE methyltransferase |
| M. ulcerans Agy99 | MUL_4762 | - | 1e-113 | 65.48% (310) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0104 | - | 1e-122 | 69.81% (308) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0095|M.smegmatis_MC2_155 ---MSSLRTDDDTWDIATSVGSTAVMVAAARAGETERDDALIRDPYAKIL
TH_4502|M.thermoresistible__bu ---MSSLRTDDDTWDIATSVGSTAVMVAASRAAETEREDALIRDPYAKVL
Mflv_0743|M.gilvum_PYR-GCK ---MSSLRTDDDTWDIASSVGATAVMVAAARAAETERHDALINDPYAKVL
Mvan_0104|M.vanbaalenii_PYR-1 ---MSSLRTDNDTWDIASSVGATAVMVAAARAAETERPEPLIDDPYAKIL
MMAR_0357|M.marinum_M ---MSTMRTHDDTWDIRTSVGATAVMVAAARAVETSKPEPLIRDPYARML
MUL_4762|M.ulcerans_Agy99 ------MRTHDDTWDIRTSVGATAVMVAAARAVETSKPEPLIRDPYARML
Mb0151|M.bovis_AF2122/97 ------MRTHDDTWDIKTSVGATAVMVAAARAVETDRPDPLIRDPYARLL
Rv0146|M.tuberculosis_H37Rv ------MRTHDDTWDIKTSVGATAVMVAAARAVETDRPDPLIRDPYARLL
MLBr_02640|M.leprae_Br4923 ------MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLL
MAV_5149|M.avium_104 ---MSSLRTHDDTWDIKSSVGTTAVMVAAARAVETEQPDPLIRDPYAKLL
MAB_4606c|M.abscessus_ATCC_199 MTGTTTARSDDDSWDIASSVGATAVMVAAARAAETRSASPLIQDPFAQLL
*:.:*:*** :***:*******:** ** ..** **:*::*
MSMEG_0095|M.smegmatis_MC2_155 VAGAGTG-VWETVLDSEFAAKIENLDPEAAAIFAHMGNYQAVRTHFFDAY
TH_4502|M.thermoresistible__bu VAGAGTG-VWELILDDDFTAKIEAADPETAAIFDHMGNYQAVRTHFFDTY
Mflv_0743|M.gilvum_PYR-GCK VEGAGAG-AWRFIADEDLVAKASESDTEVGALFEHMKNYQAVRTHFFDAF
Mvan_0104|M.vanbaalenii_PYR-1 VAGAGNG-AWQYIADDGFVAKVTESDPEIGPLFEHMKNYQAVRTHFFDAF
MMAR_0357|M.marinum_M VTNANAGVIWEAMLDQEMVAKVEAIDAETAATVEHMRSYQAVRTNFFDTY
MUL_4762|M.ulcerans_Agy99 VTNANAGVIWEAMLDQEMVAKVEAIDAETAATVEHMRSYQAVRTNFFDTY
Mb0151|M.bovis_AF2122/97 VTNAGAGAIWEAMLDPTLVAKAAAIDAETAAIVAYLRSYQAVRTNFFDTY
Rv0146|M.tuberculosis_H37Rv VTNAGAGAIWEAMLDPTLVAKAAAIDAETAAIVAYLRSYQAVRTNFFDTY
MLBr_02640|M.leprae_Br4923 VTNTGAGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTY
MAV_5149|M.avium_104 VTNSGAGVLWEAMLDPDIAARVEALDEESAAHLHHMRGYQAVRTHFFDTY
MAB_4606c|M.abscessus_ATCC_199 VERAGSG-AWSVLASDGLRQQLAELDPDADRVMQYAVDYQAVRTRFFDGF
* :. * * : . : : * : . : .******.*** :
MSMEG_0095|M.smegmatis_MC2_155 YREAADAGIRQIVILASGLDSRAYRLDWPAGTTVYEIDQPKVLEYKSATL
TH_4502|M.thermoresistible__bu FARAAEAGIRQVVILASGLDSRAYRLPWPDGTVVYEIDQPKVLEYKSATL
Mflv_0743|M.gilvum_PYR-GCK FSAAVDAGVRQIVILASGLDSRAFRLPWPAGTVVYEIDQPLVLDYKSSTL
Mvan_0104|M.vanbaalenii_PYR-1 FAAAVDAGIRQIVILASGLDSRAFRLPWPAGTTVFEIDQPLVLAYKSSTL
MMAR_0357|M.marinum_M FADAVAAGIRQVVILASGLDSRAYRLDWPAGTTVYEIDQPQVLAYKSATL
MUL_4762|M.ulcerans_Agy99 FADAVAAGIRQVVILASGLDSRAYRLDWPAGTTVYEIDQPQVLAYKSATL
Mb0151|M.bovis_AF2122/97 FASAVAAGIRQVVILASGLDSRAYRLDWPAGTIVYEIDQPKVLSYKSTTL
Rv0146|M.tuberculosis_H37Rv FASAVAAGIRQVVILASGLDSRAYRLDWPAGTIVYEIDQPKVLSYKSTTL
MLBr_02640|M.leprae_Br4923 FNNAVIDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTL
MAV_5149|M.avium_104 FADAVAAGIRQIVILASGLDSRAYRLDWPAGTTVYEIDQPQVLAYKSTTL
MAB_4606c|M.abscessus_ATCC_199 FERAAHAGITQVVILAAGLDSRAYRLDWPAGTTVYEIDQPLVLQYKRHTL
: *. *: *.****:******:** ** ** *:***** ** ** **
MSMEG_0095|M.smegmatis_MC2_155 REHGIEPVAQRREVPVDLRFDWPTALHDAGFDASLPTAWLAEGLLMYLPA
TH_4502|M.thermoresistible__bu AAHGVQPAADRREVAVDLRHDWPKALRAKGFDPQQRTAWLAEGLLMYLPA
Mflv_0743|M.gilvum_PYR-GCK AAHGVAPTAERREVPIDLRQDWPAALVATGFDPARPTAWLAEGLLMYLPA
Mvan_0104|M.vanbaalenii_PYR-1 ASHGVQPTADRREVPIDLRQDWPTALTHAGFDADQPTAWLAEGLLMYLPA
MMAR_0357|M.marinum_M AENGVTPAAERREVAIDLRQDWPSALRAAGFDPSARTAWLAEGLLMYLPA
MUL_4762|M.ulcerans_Agy99 AENGVTPAAERREVAIDLRQDWPSALRAAGFDPSARTAWLAEGLLMYLPA
Mb0151|M.bovis_AF2122/97 AENGVTPSAGRREVPADLRQDWPAALRDAGFDPTARTAWLAEGLLMYLPA
Rv0146|M.tuberculosis_H37Rv AENGVTPSAGRREVPADLRQDWPAALRDAGFDPTARTAWLAEGLLMYLPA
MLBr_02640|M.leprae_Br4923 AEHGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPA
MAV_5149|M.avium_104 AENGVTPSADRREVAVDLRQDWPAALRAAGFDPTQRTAWLAEGLLMYLPA
MAB_4606c|M.abscessus_ATCC_199 DAHDVRSACLRREVPVDLRQDWPAALQREGFDVAEPTAWLAEGLLMYLPT
:.: . . ****. *** *** ** *** *************:
MSMEG_0095|M.smegmatis_MC2_155 EAQDRLFELVTELSAPGSRIAVETAGVAATDR---REAMRERFKKFADQL
TH_4502|M.thermoresistible__bu DAQDRLFENITALSAPGSRLAVETAAEEPVDR---REQMRARMERIASQF
Mflv_0743|M.gilvum_PYR-GCK DAQDRLFAQITELSAPGSQVAAESMGVHAPDR---RERMRERFASIASQI
Mvan_0104|M.vanbaalenii_PYR-1 DAQDRLFAQITELSAPGSRVAAESMGIHAQDR---RERMRERFASITAQF
MMAR_0357|M.marinum_M EAQDRLFTQIGELSCAGSRIAAETAGNHADER---REQMRERFRKVAQTL
MUL_4762|M.ulcerans_Agy99 EAQDRLFTQIGELSCAGSRIAAETAGNHADER---REQMRERFRKVAQTL
Mb0151|M.bovis_AF2122/97 EAQDRLFTQVGAVSVAGSRIAAETAPVHGEER---RAEMRARFKKVADVL
Rv0146|M.tuberculosis_H37Rv EAQDRLFTQVGAVSVAGSRIAAETAPVHGEER---RAEMRARFKKVADVL
MLBr_02640|M.leprae_Br4923 TAQDGLFTEIGGLSAVGSRIAVETSPLHGDEW---REQMQLRFRRVSDAL
MAV_5149|M.avium_104 EAQDRLFTLIGELSPAGSRVAAETAPNHADER---RQQMRERFKKVADEI
MAB_4606c|M.abscessus_ATCC_199 EAQDRLFELIVDQSAPGSRIAVEAVGPDNDKRQEFRSKMRAHFDRARKLA
*** ** : * **::*.*: . * *: ::
MSMEG_0095|M.smegmatis_MC2_155 NLSSA-LDIQELVYDDPDRADVAEWLDAHGWRARGVHALDEMRRLDR--L
TH_4502|M.thermoresistible__bu GLEKA-LDVAQLMYDDPDRADVPEWLTGHGWRTEWVSAPDEMRRLDR--F
Mflv_0743|M.gilvum_PYR-GCK DIEP--MDITELTYEDPDRAEVAEWLAAHGWTAQAVPSQDAMRRLDR--Y
Mvan_0104|M.vanbaalenii_PYR-1 DVEP--MDITELTYEDPDRADVAQWLTAHGWRAEAVPSQDEMRRLHR--L
MMAR_0357|M.marinum_M GLEQT-IDVHELIYHDPDRALLGQWLNEHGWRANAQNACDEMHRVGR--W
MUL_4762|M.ulcerans_Agy99 GLEQT-IDVHELIYHDPDRAPLGQWLNEHGWRANAQNACDEMHRVGR--W
Mb0151|M.bovis_AF2122/97 GIEQT-IDVQELVYHDQDRASVADWLTDHGWRARSQRAPDEMRRVGR--W
Rv0146|M.tuberculosis_H37Rv GIEQT-IDVQELVYHDQDRASVADWLTDHGWRARSQRAPDEMRRVGR--W
MLBr_02640|M.leprae_Br4923 GFEQA-VDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGR--W
MAV_5149|M.avium_104 GFEQT-VDVGELMYRDDHRADVTEWLNAHGWRATAEHSTAAMRRLGR--W
MAB_4606c|M.abscessus_ATCC_199 GMDGESLDVGSLMYHDENRTDVPQWLAGRGWTVRAIPAEVEMAEAGRPGY
... :*: .* * * .*: : :** :** . : * . *
MSMEG_0095|M.smegmatis_MC2_155 VELPDDP---------DRAAFSTFVTAQRL-
TH_4502|M.thermoresistible__bu VAAAMPPGDDADAADLDDTAFSKFVTAERPS
Mflv_0743|M.gilvum_PYR-GCK VEVADSD----------GQSFSTFTTGTKL-
Mvan_0104|M.vanbaalenii_PYR-1 VEIADGD----------DQSFSTFTTAVKR-
MMAR_0357|M.marinum_M VEGVPMAD--------DKQAYSEFVTAERL-
MUL_4762|M.ulcerans_Agy99 VEGVPMAD--------DKQAYSDFVTAERL-
Mb0151|M.bovis_AF2122/97 VEGVPMAD--------DPTAFAEFVTAERL-
Rv0146|M.tuberculosis_H37Rv VEGVPMAD--------DPTAFAEFVTAERL-
MLBr_02640|M.leprae_Br4923 GDGVPMAD--------DKDAFAEFVTAHRL-
MAV_5149|M.avium_104 IENVPLAD--------DKDAFSDFVVAERR-
MAB_4606c|M.abscessus_ATCC_199 IEEDLRGN--------------TLIEAELKG
: .