For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSMASVVSRESYFDTGMDVLSDLGYGGLKLAEVCNRLGVTTGSFYHYFPNWSAYTRELIEHWREARTLRV VEAVRADPDPHRRVETLIREALALPHGAEAAIRVWSSLDPDVYTVQSVVDRLRYDILFEAAREIIGDDRN AGYFAAWGVYVIVGYEQSTLPRDGDALGWIAFQLRDALSTGRFAPSSDPTHGNTLN*
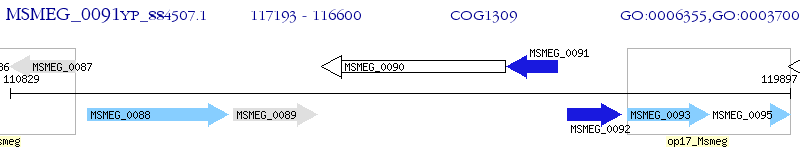
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0091 | - | - | 100% (197) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6293 | - | 5e-08 | 27.52% (109) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0745 | - | 3e-63 | 60.22% (181) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3078 | - | 1e-19 | 34.19% (155) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3423 | - | 3e-10 | 32.76% (116) | putative transcriptional regulatory protein |
| M. avium 104 | MAV_5101 | - | 1e-62 | 58.92% (185) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_1277 | - | 6e-67 | 64.61% (178) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_2701 | - | 2e-09 | 31.90% (116) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0102 | - | 8e-65 | 61.88% (181) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0091|M.smegmatis_MC2_155 ----MTSMASVVSRESYFDTGMDVLSDLGYGGLKLAEVCNRLGVTTGSFY
TH_1277|M.thermoresistible__bu -----------VTREAYFETGLAVLADLGYGGLKLAEVCNRLGVTTGSFY
Mflv_0745|M.gilvum_PYR-GCK -------MGAVSSREAFFEAGLEVLSDRGYGGLKLATVCQRLGVTTGSFY
Mvan_0102|M.vanbaalenii_PYR-1 -------MGAVASRESFFKTGLDVLADLGYGGLKLAEVCDRLGVTTGSFY
MAV_5101|M.avium_104 MPTKVPFMGNGASRSAYFEKGLEILSEVGYGGLKLAEVCLRVGVSTGSFY
MMAR_3423|M.marinum_M MAGRGTSGNRRLGVDDWVQAGFEILAEEGLSSLKLDQLCARLGVTKGSFY
MUL_2701|M.ulcerans_Agy99 MAGRSTSGNRRLGVDDWVQAGFEILAEEGLISLKLDQLCARLGVTKGSFY
MAB_3078|M.abscessus_ATCC_1997 -------MTEKVSRDDYFTAAFELLAAGGKSALTTTAVCRRIGMTTGSLY
. :. .: :*: * .*. :* *:*::.**:*
MSMEG_0091|M.smegmatis_MC2_155 HYFPNWSAYTRELIEHWREARTLRVVEAVRADPDPHRRVETLIREALALP
TH_1277|M.thermoresistible__bu HYFSGWPTYTRELIEFWRRERTTAIIESVRDTPDPRRRIGTLVQESLQLP
Mflv_0745|M.gilvum_PYR-GCK HYFANWGSYTGQLVAHWHEGLTVRVVEAVQSEADPRRRIGHLIEVGLALP
Mvan_0102|M.vanbaalenii_PYR-1 HYFPNWPAYSQELVGHWYEGMTLKVVDTVQAETDPRLRIDRLIEAGLTLP
MAV_5101|M.avium_104 HFFDNWPKYTRELIEHWQQTNTIRLAEWIEAEPDPRRRIDRIIDIGLNLP
MMAR_3423|M.marinum_M WHFADMEGYRHALVEAWAQFRDEDRDEIEGLREVEPRERLALMMALLARP
MUL_2701|M.ulcerans_Agy99 WHFADMEGYRHALVEAWAQFRDEDRDEIEGLSEVEPRERLALMMALLARP
MAB_3078|M.abscessus_ATCC_1997 HHFGSGAAFYRAVIDYWENDVTVAQRRRVDAVADPRERLATLEQAAREAD
.* . : :: * . . :
MSMEG_0091|M.smegmatis_MC2_155 --HGAEAAIRVWSSLDPDVYTVQSVVDRLRYDILFEAAREIIGDDRNAGY
TH_1277|M.thermoresistible__bu --HGAEAAIRVWSSFDPDVYEVQVAVDRQRFDIMYESALEVLGNERQAHV
Mflv_0745|M.gilvum_PYR-GCK --HGAESAIRVWSFIDPQVHEVQAAVDRQRFDVLYDSAIQILDSPRQAEL
Mvan_0102|M.vanbaalenii_PYR-1 --HSAEAAIRVWSGVDPHVGSVQEAVDRQRFDVMRESALEVLRNPRQAEL
MAV_5101|M.avium_104 --HGAEAAIRAWSSTDPEVYAVQKEVDQERHRILRDSAVQILHDDRQAQL
MMAR_3423|M.marinum_M RLWKLERAMREWARSDKTVAAAVLSADRRMVRAVQQAFCDYGFDADEAEM
MUL_2701|M.ulcerans_Agy99 RLWKLERAMREWARSDKTVAAAVLSADRRMVRAVQQAFCDYGFDADEAEM
MAB_3078|M.abscessus_ATCC_1997 --HEVEKAIRSWAMADRYVAAAQQRVDQVRHEHLTKHYIEAGFTPARADT
* *:* *: * * . .*: : . : .*
MSMEG_0091|M.smegmatis_MC2_155 FAAWGVYVIVGYEQSTLPRDGDALGWIAFQLRDALSTGRFAPSSDPTHGN
TH_1277|M.thermoresistible__bu FASLTMYILVGYEQATLPRDPGVLAWIAEQVLRTLDSGGFATIPG---GE
Mflv_0745|M.gilvum_PYR-GCK FAAWAVYLLVGYEQAILPRDISSLRWIVSQLLEALDSGRFDSVPRDE---
Mvan_0102|M.vanbaalenii_PYR-1 FAAWAVYLLVGYEQASLPADVDGLRWIVGQLLEALDSGRFTSVPGRE---
MAV_5101|M.avium_104 FADWAVYLLVGYEQATLPPDKEALQWIVGQLLDALDAGRFASAPLKKG--
MMAR_3423|M.marinum_M RANTAFAAGLGLLHMSGSALKGLFGSALKRQTAAPERNWFLEFLLRE---
MUL_2701|M.ulcerans_Agy99 RANTAFAAVLGLLHMSGSALKGLFGSALKRQTAAPERNWFLEFLLRE---
MAB_3078|M.abscessus_ATCC_1997 LARIGFSILVGAQQFGTRVDHKRLNSALDEYSAWVNSSVSAEVTTSH---
* . :* : : . . .
MSMEG_0091|M.smegmatis_MC2_155 TLN
TH_1277|M.thermoresistible__bu ---
Mflv_0745|M.gilvum_PYR-GCK ---
Mvan_0102|M.vanbaalenii_PYR-1 ---
MAV_5101|M.avium_104 ---
MMAR_3423|M.marinum_M ---
MUL_2701|M.ulcerans_Agy99 ---
MAB_3078|M.abscessus_ATCC_1997 ---