For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VCADLASAVVAFDEQTRAWQALDPKLPAAEWSPDHRAVMDDVAPVMSANADNLERLGRASDNAIVEDFTV LAAQYQRGYVEAIPTYSSADNVLWQVVASLVKAVNSGCKAS
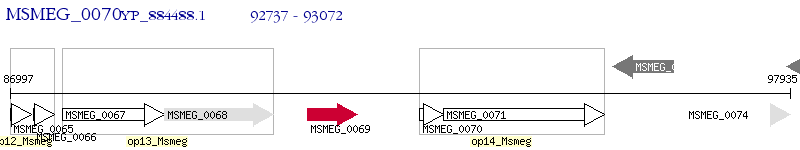
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0070 | - | - | 100% (111) | hypothetical protein MSMEG_0070 |
| M. smegmatis MC2 155 | MSMEG_4599 | - | 2e-24 | 44.44% (108) | hypothetical protein MSMEG_4599 |
| M. smegmatis MC2 155 | MSMEG_2820 | - | 3e-21 | 42.73% (110) | hypothetical protein MSMEG_2820 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0759 | - | 5e-20 | 42.59% (108) | hypothetical protein Mflv_0759 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2900 | - | 6e-22 | 42.73% (110) | PUTATIVE hypothetical protein MSMEG_2820 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0086 | - | 7e-18 | 37.61% (109) | hypothetical protein Mvan_0086 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0759|M.gilvum_PYR-GCK -----------------------------MSTPPGDRPPPPPGPPPGYP-
Mvan_0086|M.vanbaalenii_PYR-1 MKPKRRRLSALSGLTWEDGNDGARYGEVYMSSPPPDQWPAPPPPPTGPPQ
TH_2900|M.thermoresistible__bu --------------------------------------------------
MSMEG_0070|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0759|M.gilvum_PYR-GCK GGSPWGAAQPNGPRGRGAAVWVLGGLAVLVVIVLTVAATLFVTGRQSEEA
Mvan_0086|M.vanbaalenii_PYR-1 GATQWGPRTTGPTRGGAAAKWVLGGLALLVVVVITVVATILVT-QSPQTS
TH_2900|M.thermoresistible__bu --------------------------------------------------
MSMEG_0070|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0759|M.gilvum_PYR-GCK GPDPAKPSPTTAVDTSEIASADDDGPVEIITEDPTCSSWTTIVTAFVDAS
Mvan_0086|M.vanbaalenii_PYR-1 GPPVTSTPPRTTASQVAVASADDRDPAGIILDDATCQQWSPISAAFAHVA
TH_2900|M.thermoresistible__bu -----------------VASANDTGPVAIITEEPTCAPVRPILIVFSDAQ
MSMEG_0070|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0759|M.gilvum_PYR-GCK RNGWDARDPTSPAASWTPEQSRQYEAMGAAMQSIADRTVELAKRTPHRVM
Mvan_0086|M.vanbaalenii_PYR-1 SNGWNDRDPTIPASDWPPALRSQYEAVATVLNESADRSSELARKTPHRVM
TH_2900|M.thermoresistible__bu QNGWDRRDASIPASGWTHEVRVQYEEVGDAMRETADRLVPMVKLTPHRVV
MSMEG_0070|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0759|M.gilvum_PYR-GCK RILYEQFVAYSRAYVGKLSTYTEQDDRFVRVAISFSGTLNAICDSIAFGA
Mvan_0086|M.vanbaalenii_PYR-1 RELYEQFIAYSREYAERIRSYTPEANGFALVTVGLAATLTAVCDAVRSGA
TH_2900|M.thermoresistible__bu RELYEQMIAYLRAYAQAIPRYRPADDFLARSGNAAADAISNICKSIEYGA
MSMEG_0070|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0759|M.gilvum_PYR-GCK AGARAPLVPGSAPPRDLERELDPDNAKPFMTSPSSSCGQWLYATEKFTND
Mvan_0086|M.vanbaalenii_PYR-1 AGARSPLVPDAPPPSGSMAKADPDNPQRFLKDLNPVCSQWLNTAKQFGDD
TH_2900|M.thermoresistible__bu AAARARMVTPLAAPEVLAPVGDIANPQRLLTEIDPVCAEWADAVSEFQRD
MSMEG_0070|M.smegmatis_MC2_155 -----------------------------------MCADLASAVVAFDEQ
*.: :. * :
Mflv_0759|M.gilvum_PYR-GCK TAAWRGIDPNIPASELTADQRAVNAAVAPVMEAFATESQQLGRRSNNPIW
Mvan_0086|M.vanbaalenii_PYR-1 TAAWRSVDPNIPGNELDANQRAINASVMSVMEANAMETQLLGKQSSNPIW
TH_2900|M.thermoresistible__bu AAAWGTTSPDIPASQWSSEQRALNDQVMPVMQTFADRLVALGQRTSNPTL
MSMEG_0070|M.smegmatis_MC2_155 TRAWQALDPKLPAAEWSPDHRAVMDDVAPVMSANADNLERLGRASDNAIV
: ** .*.:*. : .::**: * .**.: * . **: :.*.
Mflv_0759|M.gilvum_PYR-GCK ADLAALSAQYRRAYVSALVTYSPSDNDLQVAAGSTAGAISEACRIVMD-
Mvan_0086|M.vanbaalenii_PYR-1 ADIATLSAQYRRAYVSALTTYQPADNDLQIAASSAIGTVSEACRTVGA-
TH_2900|M.thermoresistible__bu RDFAELSALYRSAYIEAIPTYMPADQYLTAASIRIAGVVNAACKALTNN
MSMEG_0070|M.smegmatis_MC2_155 EDFTVLAAQYQRGYVEAIPTYSSADNVLWQVVASLVKAVNSGCKAS---
*:: *:* *: .*:.*: ** .:*: * . .:. .*: