For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGRLVLVRHGQSVSNVERRLDTLPPGAELTPTGRVQAQAFARGGLIRPAVLVHSIATRASQTAEVIAAEL AVSAHEVVGIHEVQVGELENRCDDEAVAEFNAIYKRWHCGELDVPLPGGESANDVLARYVPVLNDLRMRY LDDEDRTGDLVVVSHGAAIRLAGAVLAGVDANFALDNHLDNAESVVLTPITDGRWSCVRWGTKTPPFCAE VDSAAGDLMASSGDPMG*
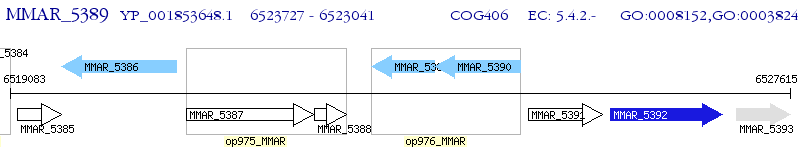
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5389 | - | - | 100% (228) | phosphoglycerate mutase |
| M. marinum M | MMAR_3747 | gpm_1 | 8e-09 | 29.05% (210) | phosphoglycerate mutase, Gpm_1 |
| M. marinum M | MMAR_1154 | lpqD | 4e-08 | 26.13% (199) | lipoprotein, LpqD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3867c | - | 1e-94 | 74.14% (232) | phosphoglycerate mutase |
| M. gilvum PYR-GCK | Mflv_1146 | - | 5e-79 | 67.66% (235) | phosphoglycerate mutase |
| M. tuberculosis H37Rv | Rv3837c | - | 1e-94 | 74.14% (232) | phosphoglycerate mutase |
| M. leprae Br4923 | MLBr_00079 | - | 2e-83 | 66.81% (232) | putative phosphoglycerate mutase |
| M. abscessus ATCC 19977 | MAB_0133 | - | 7e-73 | 60.26% (229) | phosphoglycerate mutase |
| M. avium 104 | MAV_0189 | - | 8e-89 | 71.12% (232) | phosphoglycerate mutase family protein |
| M. smegmatis MC2 155 | MSMEG_6416 | - | 1e-78 | 63.60% (228) | phosphoglycerate mutase family protein |
| M. thermoresistible (build 8) | TH_1473 | - | 1e-75 | 61.84% (228) | PROBABLE PHOSPHOGLYCERATE MUTASE (PHOSPHOGLYCEROMUTASE) |
| M. ulcerans Agy99 | MUL_5010 | - | 1e-126 | 96.49% (228) | phosphoglycerate mutase |
| M. vanbaalenii PYR-1 | Mvan_5661 | - | 1e-76 | 63.25% (234) | phosphoglycerate mutase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1146|M.gilvum_PYR-GCK -----------MTGRLVLVRHGQSHANVERRLDTRPPGAELTDLGRDQAR
Mvan_5661|M.vanbaalenii_PYR-1 -----------MSGRLVLVRHGQSHGNVARRLDTRPPGAELTDLGREQAR
MMAR_5389|M.marinum_M -----------MSGRLVLVRHGQSVSNVERRLDTLPPGAELTPTGRVQAQ
MUL_5010|M.ulcerans_Agy99 -----------MSGRLVLVRHGQSVSNVERRLDTLPPGAELTPTGRAQAQ
Mb3867c|M.bovis_AF2122/97 -----------MSGRLVLLRHGQSYGNVERRLDTLPPGTALTPLGRDQAR
Rv3837c|M.tuberculosis_H37Rv -----------MSGRLVLLRHGQSYGNVERRLDTLPPGTALTPLGRDQAR
MLBr_00079|M.leprae_Br4923 -----------MSSRLVLLRHGQSYGNVEGRLDTRPPGASLTPFGRDQAR
MAV_0189|M.avium_104 -----------MSGRLILVRHGQSYGNVERRLDTRPPGAELTPLGRDQAR
MSMEG_6416|M.smegmatis_MC2_155 -----------MSGRLVLVRHGQSRANVDRRLDTRPPGAELTDLGHDQAR
MAB_0133|M.abscessus_ATCC_1997 MARRDAGREAAMSGRLVLVRHGQSYGNVERRLDTKPPGAALTELGLQQAR
TH_1473|M.thermoresistible__bu -----------MSGRLVLVRHGQSQANVERRLDTAPPGAELTDLGRQQAR
*:.**:*:***** .** **** ***: ** * **:
Mflv_1146|M.gilvum_PYR-GCK TFARTLAR-PAAIVTHSIATRAVQTAGHIAAEFGGALS--AGAMAFEGLH
Mvan_5661|M.vanbaalenii_PYR-1 TFARELTR-PPAMVTHSIATRAVQTAGHIYSELSGVIRSQSGTLEFEGLH
MMAR_5389|M.marinum_M AFARGGLI-RPAVLVHSIATRASQTAEVIAAELA------VSAHEVVGIH
MUL_5010|M.ulcerans_Agy99 AFARGGLI-RPAMLVHSIATRASQAAEVVAAVLT------VSAHEVVGIH
Mb3867c|M.bovis_AF2122/97 AFARSGCR-RPALLAHSVAIRAYQTAAVVAAELD------MVAHEVAGIH
Rv3837c|M.tuberculosis_H37Rv AFARSGCR-RPALLAHSVAIRAYQTAAVVAAELD------MVAHEVAGIH
MLBr_00079|M.leprae_Br4923 VFAQAAG--RPVLLVHSVAIRALETAAVLGAEFD------VPIREIAGIH
MAV_0189|M.avium_104 EFARRSG--RPAMLAHSVATRASQTAAVIGGQLA------LPPVELGGIH
MSMEG_6416|M.smegmatis_MC2_155 RVVTAFPR-RPALVAHSVARRAAQTASGIAGEIG------LTPQEFDGVH
MAB_0133|M.abscessus_ATCC_1997 LFAKRYEEHAPAVLVHSVAVRAVQTAAGIAEHLG------VQAEQIEGLH
TH_1473|M.thermoresistible__bu AFARQLRR-PPGLLVHSVAVRAEQTAGEAGSHLG------QQAHVFEGIH
.. . ::.**:* ** ::* : . *:*
Mflv_1146|M.gilvum_PYR-GCK EVQVGELEDRTDEAAHDEFNAVYRRWHGGELDVALPGGETAQQVLDRYVP
Mvan_5661|M.vanbaalenii_PYR-1 EVQVGHLEDRCDEAAHDEFNAIYQRWHGGELDLAMPGGESAQQVLDRYVP
MMAR_5389|M.marinum_M EVQVGELENRCDDEAVAEFNAIYKRWHCGELDVPLPGGESANDVLARYVP
MUL_5010|M.ulcerans_Agy99 EVQVGELENRCDDEAAAEFNAIYKRWHCGELDVPLPGGESANDVLARYVP
Mb3867c|M.bovis_AF2122/97 EVQVGELENRNDDEAVAEFNATYSRWHRGELDVPLPGGETANDVLDRYLP
Rv3837c|M.tuberculosis_H37Rv EVQVGELENRNDDEAVAEFNATYSRWHRGELDVPLPGGETANDVLDRYLP
MLBr_00079|M.leprae_Br4923 EVQVGELENRNDDDAIAEFNAIYDRWHHGELDVPLPGGETANDVLDRYLP
MAV_0189|M.avium_104 EVQVGRLENRNDDEAVAEFNAIYERWHRGELEVPLPGGETGSEVLDRYLP
MSMEG_6416|M.smegmatis_MC2_155 EVQVGELEDRNDDVAIAQFEAIYQRWHQGELEVPMPGGESAVDVLDRYVP
MAB_0133|M.abscessus_ATCC_1997 EVQAGELEDRTDLEAFAVFDRTYERWHFGDLDARMPGGESAQDVFDRYLP
TH_1473|M.thermoresistible__bu EVQAGELENRNDDEAITEFETVYQQWHQGNLKVNMPGGENGVEVLDRYVS
***.*.**:* * * *: * :** *:*. :****.. :*: **:.
Mflv_1146|M.gilvum_PYR-GCK VLDQLRMRYLDDETWHGDIVVVSHGAAIRLVSALLAGVDSRFAVDHHLAN
Mvan_5661|M.vanbaalenii_PYR-1 VLEQLRMRYLEDGSWHGDIVVVSHGAAIRLVAAVLAGVDSGFAVDHHLAN
MMAR_5389|M.marinum_M VLNDLRMRYLDDEDRTGDLVVVSHGAAIRLAGAVLAGVDANFALDNHLDN
MUL_5010|M.ulcerans_Agy99 VLNDLRMRYLDDEDRTGDLVVVSHGAAIRPAGAVLAGVDANFALDNHLDN
Mb3867c|M.bovis_AF2122/97 VLADLRMRYLDDGDWDGDIVVVSHSAAIRLAAAVLAGVDGNFVLDNHLEN
Rv3837c|M.tuberculosis_H37Rv VLADLRMRYLDDGDWDGDIVVVSHSAAIRLAAAVLAGVDGNFVLDNHLEN
MLBr_00079|M.leprae_Br4923 VLVDLRMRYLDDGDWTGDVVVVSHGAAIRLVSAVLAEVDGRFALENRVNN
MAV_0189|M.avium_104 VLTELRLRYLDDHDFHGDIVVVSHGAAIRLAAAVLAGVEADFALDHHLDN
MSMEG_6416|M.smegmatis_MC2_155 VVDHLRLNYLDDHDFDSDIVLVSHGAAIRLVAAVLAGVDGSFALEHHLAN
MAB_0133|M.abscessus_ATCC_1997 AVADLRLRHLENDASTGDVVIVSHGAAIRLVAAALAGVDPGFAVNRHLRN
TH_1473|M.thermoresistible__bu VITQLRLHYLDNDAWTDDIVVVSHGAAIRLVAATLAGVDSTFALDRHLGN
.: .**:.:*:: .*:*:***.**** ..* ** *: *.::.:: *
Mflv_1146|M.gilvum_PYR-GCK TESVVLAPITDGRWSCVQWGKLTPPFVGDAPVTTTAG---DAPRSADPMG
Mvan_5661|M.vanbaalenii_PYR-1 TESVVLAPVTEGRWSCVQWGKLTPPFGPEAPVVTSP----EAAQSADPMG
MMAR_5389|M.marinum_M AESVVLTPITDGRWSCVRWGTKTPPFCAEVDSAAG----DLMASSGDPMG
MUL_5010|M.ulcerans_Agy99 AESVVLTPITDGRWSCVRWGTKTPPFCAEVDSAAG----DLMASSGDPMG
Mb3867c|M.bovis_AF2122/97 VESVVLAPITDGRWSCVQWGLRKPPFCPDPAEAAASPVTHAVTSSTDPMG
Rv3837c|M.tuberculosis_H37Rv VESVVLAPITDGRWSCVQWGLRKPPFCPDPAEAAASPVTHAVTSSTDPMG
MLBr_00079|M.leprae_Br4923 AESVVLVPVTDGRWSCVRWGALTPPFYPHLHRPDSTLVTDVVMSGRDSLG
MAV_0189|M.avium_104 AQSVALTPITDGRWSCVRWGTLTPPFYPEAAEIPATPVADAVRSTTDPMG
MSMEG_6416|M.smegmatis_MC2_155 TESVILTPTTDGRWSCVRWATLTPPFHPEPDVQPVT---DALTS-KDPMG
MAB_0133|M.abscessus_ATCC_1997 AEAVVLAPVTDGRWSCVHWGDAVAPF-PHQEAASA----EELAQSADPMG
TH_1473|M.thermoresistible__bu TESVVLAPITDGRWSCLQWGKESPPFYPEPDVQPV----EDALHSVDPMG
.::* *.* *:*****::*. .** . *.:*