For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RGVSVVIRLLCVAVLSVTLGGITVATGSTGKANAAPYETLMVPSGAMGRDIPVAFLAGGPHAAYLLDAFN AGPDVSNWVTAGNAMNTLGGKGISVVAPAGGAFSMYTDWEQDGSKQWDTFLSSELPDWLAANKGLAPGGH AAVGASQGGYGAMALAAFHPDRFGFAGSLSGFLYPSSTTTNGAILAGMQQFGGVDGNGMWGAPQLGRWKW HDPWVHAALLAQNNTRVWVWSPTNPGASDPAAMIGQAGAAMGDSRSFYQQYRNVGGHNGHFDFPGGGDNG WGSWSAQLGAMSGDIVGAIR*
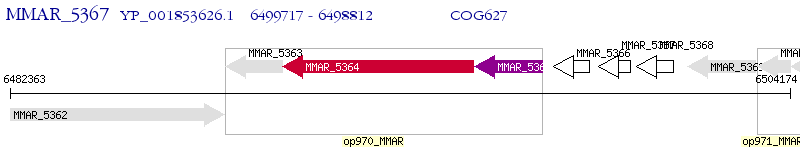
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5367 | fbpD | - | 100% (301) | secreted Mpt51/Mpb51 antigen protein FbpD |
| M. marinum M | MMAR_0328 | fbpC | 1e-57 | 40.52% (306) | secreted antigen 85-C FbpC |
| M. marinum M | MMAR_2777 | fbpB | 1e-51 | 38.26% (311) | secreted antigen 85-B FbpB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3833c | fbpD | 1e-156 | 85.38% (301) | secreted MPT51/MPB51 antigen protein FBPD (MPT51/MPB51 |
| M. gilvum PYR-GCK | Mflv_1165 | - | 1e-113 | 64.12% (301) | putative esterase |
| M. tuberculosis H37Rv | Rv3803c | fbpD | 1e-156 | 85.38% (301) | secreted MPT51/MPB51 antigen protein FBPD (MPT51/MPB51 |
| M. leprae Br4923 | MLBr_00098 | fbpC | 1e-143 | 78.81% (302) | antigen 85C, mycolyltransferase |
| M. abscessus ATCC 19977 | MAB_1579 | - | 2e-55 | 39.50% (319) | antigen 85-B precursor |
| M. avium 104 | MAV_0215 | - | 1e-155 | 85.71% (301) | antigen 85-C |
| M. smegmatis MC2 155 | MSMEG_6396 | - | 1e-115 | 64.67% (300) | antigen 85-C |
| M. thermoresistible (build 8) | TH_2468 | - | 1e-120 | 67.55% (302) | PUTATIVE putative esterase |
| M. ulcerans Agy99 | MUL_4986 | fbpD | 0.0 | 99.34% (301) | secreted Mpt51/Mpb51 antigen protein FbpD |
| M. vanbaalenii PYR-1 | Mvan_5644 | - | 1e-114 | 65.64% (291) | putative esterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1165|M.gilvum_PYR-GCK -----MRGMVRAVAAVAMAALFW--SGAVAPFTPATPVARAQG-----VE
Mvan_5644|M.vanbaalenii_PYR-1 -----MHAMLRAIAALTVAV-----AAVLAPVT--APTAGAQG-----VE
MSMEG_6396|M.smegmatis_MC2_155 -----MRRGLSLVRALMLTVVLA--AGLWTVSATSGAPARADG-----VE
TH_2468|M.thermoresistible__bu -----MN-LLSVFFRMMCAAVLA--AVGVTVST--ATATAQPA-----VE
MMAR_5367|M.marinum_M -----MRGVSVVIRLLCVAVLSVT-LGGITVATGSTGKANAAP-----YE
MUL_4986|M.ulcerans_Agy99 -MEVDMRGVSVVTRLLCVAVLSVT-LGGITVATGSTGKANAAP-----YE
Mb3833c|M.bovis_AF2122/97 -----MKGRSALLRALWIAALSFG-LGGVAVAAEPT--AKAAP-----YE
Rv3803c|M.tuberculosis_H37Rv -----MKGRSALLRALWIAALSFG-LGGVAVAAEPT--AKAAP-----YE
MAV_0215|M.avium_104 MDMTVVRGVSALLRVFCIAVLAAG-LGVALQPAAVTGAARAAG-----YE
MLBr_00098|M.leprae_Br4923 -----MRGLSAVVRVLCVAALAVGVFAAAVLLAGTAGNAKAAG-----YE
MAB_1579|M.abscessus_ATCC_1997 -MEFLQKMRGATARLAAVAAVAAAILPGLIGVTGGSAIAGAFSRPGLPVE
. :. : : *
Mflv_1165|M.gilvum_PYR-GCK MLMVPSAAMGRAIPVAFQGGGPHAVVLLDAFNAAPDVSNWVTAGNAMNTL
Mvan_5644|M.vanbaalenii_PYR-1 MLMVPSAAMGRSIPVAFQGGGPHAVVLLDAFNAAPDVSNWVTAGNAMGTL
MSMEG_6396|M.smegmatis_MC2_155 YLMVPSAAMGRDIPVAFQAGGPHAVFLLDAFNAAPDVSNWVNAGSAMSTL
TH_2468|M.thermoresistible__bu QLMVPSAAMGRDIPVSFMAGGPHAVVLLDAFNAAPDVSNWVTVGNAMNTL
MMAR_5367|M.marinum_M TLMVPSGAMGRDIPVAFLAGGPHAAYLLDAFNAGPDVSNWVTAGNAMNTL
MUL_4986|M.ulcerans_Agy99 TLMVPSGAMGRDIPVAFLAGGPHAAYLLDAFNAGPDVSNWVTAGNAMNTL
Mb3833c|M.bovis_AF2122/97 NLMVPSPSMGRDIPVAFLAGGPHAVYLLDAFNAGPDVSNWVTAGNAMNTL
Rv3803c|M.tuberculosis_H37Rv NLMVPSPSMGRDIPVAFLAGGPHAVYLLDAFNAGPDVSNWVTAGNAMNTL
MAV_0215|M.avium_104 SLMVPSAAMGRDIPVAFLAGGPHAVYLLDAFNAAPDVSNWVTAGNAMNTL
MLBr_00098|M.leprae_Br4923 SLMVPSNAMGRDIPVAFMAGGPHAVYLLDAFNAALDVSNWVTAGNAMTTL
MAB_1579|M.abscessus_ATCC_1997 YLQVPSPAMGRDIVVEFQPGGPHAVYLLDGQRAREDYNGWDIETTAFEDY
* *** :*** * * * *****. ***. .* * ..* .*:
Mflv_1165|M.gilvum_PYR-GCK SGLGISVAAPAGGAWSMYTNWEQDG-------SKQWETFLADELPNWLAA
Mvan_5644|M.vanbaalenii_PYR-1 SGRGISVAAPAGGAWSMYTNWEQDG-------SKQWETFLADELPNWLAA
MSMEG_6396|M.smegmatis_MC2_155 AGRGISVAAPAGGAWSLYTNWEQDG-------SKQWETFLTAELPGWLAA
TH_2468|M.thermoresistible__bu AGKGISVVAPAGGAWSLYTNWENDG-------SKQWETFLANELPDWLAA
MMAR_5367|M.marinum_M GGKGISVVAPAGGAFSMYTDWEQDG-------SKQWDTFLSSELPDWLAA
MUL_4986|M.ulcerans_Agy99 GGKGISVVAPAGGAFSMYTDWEQDG-------SKQWDTFLSSELPDWLAA
Mb3833c|M.bovis_AF2122/97 AGKGISVVAPAGGAYSMYTNWEQDG-------SKQWDTFLSAELPDWLAA
Rv3803c|M.tuberculosis_H37Rv AGKGISVVAPAGGAYSMYTNWEQDG-------SKQWDTFLSAELPDWLAA
MAV_0215|M.avium_104 GGKGISVVAPAGGAWSMYTNWEQDG-------SKQWDTFLSSELPDWLAA
MLBr_00098|M.leprae_Br4923 GGRGISVVAPAGGAYSMYTNWENDG-------SKQWDTFLSSELPDWLAT
MAB_1579|M.abscessus_ATCC_1997 YQSGISVVMPVGGQSSNYTDWYNPAKGKDGVWTYKWETFLTTELPQWLGV
****. *.** * **:* : . : :*:***: *** **..
Mflv_1165|M.gilvum_PYR-GCK NKGLAPGGHGIVGAAMGGTGALTMATFHPDRYRFAGSLSGFLTPSATAMN
Mvan_5644|M.vanbaalenii_PYR-1 NKGLAPNGHAIVGAAQGGTGALTMATFHPDRYRFAGSLSGFLTPSATALN
MSMEG_6396|M.smegmatis_MC2_155 NKGLAPDGHAVVGAAQGGTAAVTLAAFHPGMFRFAGSLSGFLTPSATTLN
TH_2468|M.thermoresistible__bu NKGLAPGGHGIVGASQGGTAALTLAAFYPNRYRYAGSMSGFLTPSATALN
MMAR_5367|M.marinum_M NKGLAPGGHAAVGASQGGYGAMALAAFHPDRFGFAGSLSGFLYPSSTTTN
MUL_4986|M.ulcerans_Agy99 NKGLAPGGHAAVGASQGGYGAMALAAFHPDRFGFAGSLSGFLYPSSTTTN
Mb3833c|M.bovis_AF2122/97 NRGLAPGGHAAVGAAQGGYGAMALAAFHPDRFGFAGSMSGFLYPSNTTTN
Rv3803c|M.tuberculosis_H37Rv NRGLAPGGHAAVGAAQGGYGAMALAAFHPDRFGFAGSMSGFLYPSNTTTN
MAV_0215|M.avium_104 NKGLAPGGHAAVGASQGGYGAMALAAFHPDRFGFAGSLSGFLYPSSTNYN
MLBr_00098|M.leprae_Br4923 KRGLAPDGHAAVGASQGGYAALALAAFHPDRFGFAGSLSGFVYPSSTNYN
MAB_1579|M.abscessus_ATCC_1997 NKGISPAANAVVGLSMAGPSALTLAIYHPQQFVYAGALSAPLHPSDQKWQ
::*::* .:. ** : .* .*:::* ::* : :**::*. : ** :
Mflv_1165|M.gilvum_PYR-GCK GAITDGLARFGGLDIRNMWGLPQLGRWKWHDPDVHSQLLANNNTRLWIYS
Mvan_5644|M.vanbaalenii_PYR-1 GAITAGLARFGGVDTRNMWGLPQLGRWKWHDPDVHAQLLVNNNTRLWIYS
MSMEG_6396|M.smegmatis_MC2_155 GAITAGLARFGNVDANRMWGPPQFGRWKWHDPAVHVQLLADSNTRLWVYS
TH_2468|M.thermoresistible__bu GAITAGLQRFGGVETTNMWGLPQAGRWKWRDPDVHAQLLVNNNTRLWVFS
MMAR_5367|M.marinum_M GAILAGMQQFGGVDGNGMWGAPQLGRWKWHDPWVHAALLAQNNTRVWVWS
MUL_4986|M.ulcerans_Agy99 GAILAGMQQFGGVDGNGMWGAPQLGRWKWHDPWVHAALLAQNNTRVWVWS
Mb3833c|M.bovis_AF2122/97 GAIAAGMQQFGGVDTNGMWGAPQLGRWKWHDPWVHASLLAQNNTRVWVWS
Rv3803c|M.tuberculosis_H37Rv GAIAAGMQQFGGVDTNGMWGAPQLGRWKWHDPWVHASLLAQNNTRVWVWS
MAV_0215|M.avium_104 GAILAGLQQYGGVDGNGMWGAPQLGRWKWHDPYVHASLLAQNNTRVWVWS
MLBr_00098|M.leprae_Br4923 GAILAGLQQFGGIDGNGMWGAPQLGRWKWHDPYVHASLLAQNNTRVWVYS
MAB_1579|M.abscessus_ATCC_1997 --IRVAMSDAGGFNADDMWGPDSDPAWARNDPFLHIDQLIANNTRLWIYC
* .: *..: *** . * .** :* * .***:*::.
Mflv_1165|M.gilvum_PYR-GCK PG------TTTCTDVPAMIG--YCDQAQGSNRTFYQHYRAVGGSNGHFDF
Mvan_5644|M.vanbaalenii_PYR-1 PA------TTTCTDVPAMIG--YCDQAQGSNRTFYQHYRGIGGGNGHFDF
MSMEG_6396|M.smegmatis_MC2_155 PG------TLTCSDPAAMIG--YCDQAQGSNRQFYSDYRRVGGSNGHFDF
TH_2468|M.thermoresistible__bu PQ------TLTAADPAAMIG--YADQAQGSNRTFYAHYRQIGGRNGHFDF
MMAR_5367|M.marinum_M PT------NPGASDPAAMIG--QAGAAMGDSRSFYQQYRNVGGHNGHFDF
MUL_4986|M.ulcerans_Agy99 PT------NPGASDPAAMIG--QAGAAMGDSRSFYQQYRNVDGHNGHFDF
Mb3833c|M.bovis_AF2122/97 PT------NPGASDPAAMIG--QAAEAMGNSRMFYNQYRSVGGHNGHFDF
Rv3803c|M.tuberculosis_H37Rv PT------NPGASDPAAMIG--QAAEAMGNSRMFYNQYRSVGGHNGHFDF
MAV_0215|M.avium_104 PT------NMGGDD-AAMIG--QAGAAMGSSREFYQQYRSNGGHNGHFDF
MLBr_00098|M.leprae_Br4923 PM------TMG-GDIDAMIG--QAVASMGSSREFYQQYRSVGGHNGHFDF
MAB_1579|M.abscessus_ATCC_1997 GSGDATDLDKDRNGLEVISGGVIEGQVIDSDKKFAEAYGSAGGANAHFEF
. .: * ...: * * .* *.**:*
Mflv_1165|M.gilvum_PYR-GCK PTSGNHDWGSWSGQLAAMTGELVATIR-------
Mvan_5644|M.vanbaalenii_PYR-1 PTSGNHDWGSWSGQLAAMSGELVATIG-------
MSMEG_6396|M.smegmatis_MC2_155 PASGQHDWGSWAPQLAYMSGELVATIK-------
TH_2468|M.thermoresistible__bu PTSGDHGWSSWGPELGKMSGDLVAAIR-------
MMAR_5367|M.marinum_M PGGGDNGWGSWSAQLGAMSGDIVGAIR-------
MUL_4986|M.ulcerans_Agy99 PGGGDNGWGSWSAQLGAMSGDIVGAIR-------
Mb3833c|M.bovis_AF2122/97 PASGDNGWGSWAPQLGAMSGDIVGAIR-------
Rv3803c|M.tuberculosis_H37Rv PASGDNGWGSWAPQLGAMSGDIVGAIR-------
MAV_0215|M.avium_104 PGGGDNGWGSWAGQLGAMSGDIVGAIR-------
MLBr_00098|M.leprae_Br4923 SGGGDNGWGAWAPQLAAMSGDIVGAIR-------
MAB_1579|M.abscessus_ATCC_1997 PKGGIHNWTYWGNQLRAMKADLVGYLTKPGTAAV
. .* :.* *. :* *..::*. :