For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HYLMTHLGTAWALTMVHLRLSLIPVLIGTAIAIPLGLFVQHAPTLRRLTTATASVVFTIPSLALFVVLPL IIKTRILQEANVIIALSAYTTALLVRAVLEALDAVPAQVRDAAVAVGYSPIARMLKVELPLSIPVLVSGL RVVVVTNIAMVSVGAVIGIGGLGTWFTAGYLMDKSDQIVAGITVMFVLAILIDMVLNVAGRLATPWERSS RGRRRFRSPIVGGAR*
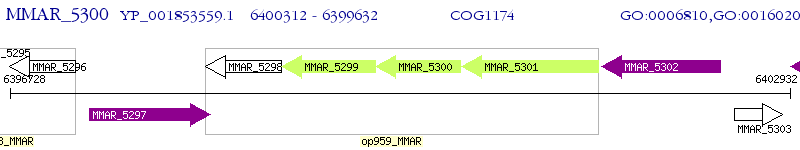
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5300 | proW | - | 100% (226) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) transport integral membrane protein ABC transporter ProW |
| M. marinum M | MMAR_0035 | - | 3e-18 | 28.37% (208) | ABC transporter permease |
| M. marinum M | MMAR_0033 | - | 3e-17 | 26.04% (192) | hypothetical protein MMAR_0033 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 3e-98 | 79.91% (229) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_1209 | - | 2e-78 | 68.57% (210) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3757c | proW | 3e-98 | 79.91% (229) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 9e-08 | 27.70% (148) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_0263 | - | 6e-81 | 68.40% (212) | osmoprotectant ABC transporter ProW |
| M. avium 104 | MAV_0319 | - | 4e-85 | 70.18% (228) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_6332 | - | 6e-81 | 70.00% (210) | amino acid ABC transporter, permease protein |
| M. thermoresistible (build 8) | TH_0698 | - | 1e-17 | 28.43% (197) | ABC transporter, permease protein OpuCB |
| M. ulcerans Agy99 | MUL_4375 | proW | 1e-120 | 99.56% (226) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_5599 | - | 5e-79 | 68.57% (210) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1209|M.gilvum_PYR-GCK ----------------------------------MRYLFTHLDDLWVLTL
Mvan_5599|M.vanbaalenii_PYR-1 ----------------------------------MRYLVTHLDALWTLSL
MSMEG_6332|M.smegmatis_MC2_155 ----------------------------------MQYLLTHLDDLWELTV
MAB_0263|M.abscessus_ATCC_1997 ----------------------------------MRYLFTHLGDAWQLSL
MMAR_5300|M.marinum_M ----------------------------------MHYLMTHLGTAWALTM
MUL_4375|M.ulcerans_Agy99 ----------------------------------MHYLMTHLGTAWALTM
Mb3783c|M.bovis_AF2122/97 ----------------------------------MHYLMTHPGAAWALTV
Rv3757c|M.tuberculosis_H37Rv ----------------------------------MHYLMTHPGAAWALTV
MAV_0319|M.avium_104 ----------------------------------MHYLLTHLDDAARLTV
TH_0698|M.thermoresistible__bu -------------------------------MDFIEFVRERWSVLSFLAY
MLBr_02093|M.leprae_Br4923 MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPLVWLLSV
:: *:
Mflv_1209|M.gilvum_PYR-GCK IHLR--------------------------------LSLIPIVLGLLIAV
Mvan_5599|M.vanbaalenii_PYR-1 IHLR--------------------------------LSLIPILLGLLIAV
MSMEG_6332|M.smegmatis_MC2_155 IHLR--------------------------------LSLVPIVLGLLIAV
MAB_0263|M.abscessus_ATCC_1997 IHLR--------------------------------LSLVPILIGLAIAI
MMAR_5300|M.marinum_M VHLR--------------------------------LSLIPVLIGTAIAI
MUL_4375|M.ulcerans_Agy99 VHLR--------------------------------LSLIPVLIGTAIAI
Mb3783c|M.bovis_AF2122/97 VHLR--------------------------------LSLLPVLIGLMSAV
Rv3757c|M.tuberculosis_H37Rv VHLR--------------------------------LSLLPVLIGLMSAV
MAV_0319|M.avium_104 VHLR--------------------------------LSLVPVLIGLAIAL
TH_0698|M.thermoresistible__bu QHMS--------------------------------LVVQTLLLSTLLAV
MLBr_02093|M.leprae_Br4923 VVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVVAAIMAV
: ::. *:
Mflv_1209|M.gilvum_PYR-GCK PLG------ALVQRTTALRRLTTVTASIIFTIPSLALFVVLPLIIPTRI-
Mvan_5599|M.vanbaalenii_PYR-1 PLG------ALVQRTTVLRRLTTVTASIVFTIPSLALFVVLPLIIPTRI-
MSMEG_6332|M.smegmatis_MC2_155 PLG------ALVQRTTTLRRLTTVAASIIFTIPSLALFVALPLIIPTRI-
MAB_0263|M.abscessus_ATCC_1997 PCG------ALLHRRRTVRRVTTVIASIVFTIPSLALFVALPLIIPTRI-
MMAR_5300|M.marinum_M PLG------LFVQHAPTLRRLTTATASVVFTIPSLALFVVLPLIIKTRI-
MUL_4375|M.ulcerans_Agy99 PLG------LFVQHAPTLRRLTTATASVVFTIPSLALFVVLPLIIKMRI-
Mb3783c|M.bovis_AF2122/97 PLG------LLVQRAPLLRRLTTATASVIFTIPSLALFVVLPLIIGTRI-
Rv3757c|M.tuberculosis_H37Rv PLG------LLVQRAPLLRRLTTATASVIFTIPSLALFVVLPLIIGTRI-
MAV_0319|M.avium_104 PLG------MLVQRRRFARRLTTAVASVVFTIPSLALFVVLPMIIGTRI-
TH_0698|M.thermoresistible__bu AVG------VMLYRSTWGRSLGDSVTSLGLTVPSYALLGVLVGIFGVGV-
MLBr_02093|M.leprae_Br4923 PLGSMTAVYLVEYGSGPFVRVTTFMVDVLAGVPSIVAALFIFCLWIATLG
. * . : ..: :** . : : :
Mflv_1209|M.gilvum_PYR-GCK --LDEANVIVALTLYTVALLVRAVPEALDAVSPAVLDAATAVGYRPLTRM
Mvan_5599|M.vanbaalenii_PYR-1 --LDEANVIVALTLYTVALLVRAVPEALDAVSPAVLDAATAVGYRPLTRM
MSMEG_6332|M.smegmatis_MC2_155 --LDEANVIVALTLYTTALLVRAVPEALDAVPEHVRDAATAVGYRPLTRM
MAB_0263|M.abscessus_ATCC_1997 --LDEANVMVALTLYTTALLIRAVPEALDAIAPQVRDAATAVGYRPLARL
MMAR_5300|M.marinum_M --LQEANVIIALSAYTTALLVRAVLEALDAVPAQVRDAAVAVGYSPIARM
MUL_4375|M.ulcerans_Agy99 --LQEANVIIALSAYTTALLVRAVLEALDAVPAQVRDAAVAVGYSPIARM
Mb3783c|M.bovis_AF2122/97 --LDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQM
Rv3757c|M.tuberculosis_H37Rv --LDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQM
MAV_0319|M.avium_104 --LDEANVMVALTAYTAALLVRAVLEALDAVPAQISDAATAVGYPRLTRM
TH_0698|M.thermoresistible__bu --LPS---VVLLTFFGFLPILRNVLVGLNGVDAGLVESARAMGMSRPATL
MLBr_02093|M.leprae_Br4923 FQQSAFAVSLALVLLMLPVVVRSTEEILRLVPDELREACYALGIPKWKTI
: * ::* . * : : ::. *:* :
Mflv_1209|M.gilvum_PYR-GCK LKVELPLALPVLIASLRVVAVTNISMVAVGSVIG--IGGLGTWFTEGYQ-
Mvan_5599|M.vanbaalenii_PYR-1 LKIELPLALPVLIASLRVVAVTNISMVAVGSVIG--IGGLGTWFTEGYQ-
MSMEG_6332|M.smegmatis_MC2_155 LKVELPLSIPVLVASLRVVAVTNISMVSVGSVIG--IGGLGTWFTEGYQ-
MAB_0263|M.abscessus_ATCC_1997 VRVELPLSIPVLVAGLRVVAVTNISMVSVGSVIG--IGGLGTWFTEGYQ-
MMAR_5300|M.marinum_M LKVELPLSIPVLVSGLRVVVVTNIAMVSVGAVIG--IGGLGTWFTAGYL-
MUL_4375|M.ulcerans_Agy99 LKVELPLSIPVLVSGLRVVVVTNIAMVSVGAVIG--IGGLGTWFTAGYL-
Mb3783c|M.bovis_AF2122/97 LKVELPLSIPVLVAGLRVVAVTNIAMVSVGSVIG--IGGLGTWFTAGYQ-
Rv3757c|M.tuberculosis_H37Rv LKVELPLSIPVLVAGLRVVAVTNIAMVSVGSVIG--IGGLGTWFTAGYQ-
MAV_0319|M.avium_104 MKVELPLAVPVLIAGLRVVVVTNIAMVSVGSVIG--IGGLGSWFTQGYQ-
TH_0698|M.thermoresistible__bu LRLELPLAWPVIMTGVRISAQMLMGIAAIAAFAL--GPGLGGYIFSGISR
MLBr_02093|M.leprae_Br4923 VRIVFPIATPGIISGVLLSIARVIGETAPVLVLVGYSRSINFDIFDGNMA
::: :*:: * :::.: : :. .: . .:. : *
Mflv_1209|M.gilvum_PYR-GCK -------ANKSDQIIAGIVAIFVLAIVIDTLIMWLGKLLTPWTRAQPRST
Mvan_5599|M.vanbaalenii_PYR-1 -------ANKSDQIIAGIVAIFVLAIVIDTLIMLAGKALTPWTRAQPRAT
MSMEG_6332|M.smegmatis_MC2_155 -------SNKSSQIIAGIIAIFVLAIVVDTLIMLAGKLIAPWNRAEVTA-
MAB_0263|M.abscessus_ATCC_1997 -------ANKSSQIIAGIVAIFLLAVIVDSLLVVLGRVATPWARATKTAG
MMAR_5300|M.marinum_M -------MDKSDQIVAGITVMFVLAILIDMVLNVAGRLATPWERSSRGRR
MUL_4375|M.ulcerans_Agy99 -------MDKSDQIVAGITVMFVLAILIDMVLNVAGRLATPWERSSRGRR
Mb3783c|M.bovis_AF2122/97 -------TNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPRAAR
Rv3757c|M.tuberculosis_H37Rv -------TNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPRAAR
MAV_0319|M.avium_104 -------TGKSDQILAGIIALFALAVAVDGLIVLGGRLVTPWAHAVRGGA
TH_0698|M.thermoresistible__bu MG----GANATNSIIVATVGILLLAVILDAVLNLVTRLTTPRGIRV----
MLBr_02093|M.leprae_Br4923 SLPLLIYTELTNPEYAGFLRVWGAALTLIILVAGINLFGAAFRFLATRRC
:. .. : *: : :: :.
Mflv_1209|M.gilvum_PYR-GCK RTQKAGASA-----
Mvan_5599|M.vanbaalenii_PYR-1 RAPRAEAGAPT---
MSMEG_6332|M.smegmatis_MC2_155 --------------
MAB_0263|M.abscessus_ATCC_1997 AT------------
MMAR_5300|M.marinum_M RFR---SPIVGGAR
MUL_4375|M.ulcerans_Agy99 RFR---SPIVGGAR
Mb3783c|M.bovis_AF2122/97 RRRQVAAPITGGAR
Rv3757c|M.tuberculosis_H37Rv RRRQVAAPITGGAR
MAV_0319|M.avium_104 RRS-VVAPVVGGAR
TH_0698|M.thermoresistible__bu --------------
MLBr_02093|M.leprae_Br4923 SGCR----------