For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSQTPATTRTAFGGISSRAWEHPADRTALSALRRLKGFDQILKLLSGMLRERQHRLLYLASAARVGPRQF ADLDALLDECVDVLDAPTKPEVYVTQSPMVNAYTIGMDAPFIVITSGLYDLMTHDELRFVVGHELGHALS GHATYRTMLMHLMRLAGSLGFLPIGGWALRAIVAALLEWQRKSELSGDRAGLLCCQDLDTAIRVEMKLAG GSRLDKLDSEAFLAQAAEYERSGDMRDGLLKLLNLELQTHPFSVLRVAELTKWVDAGGYGKVMSGSYPLR LDDGQSSIADDLGEAARHYKDGFDQSDDPLIKGIRDGLGGIVDGVGRAATSAADSLGRKINEWRHNPK
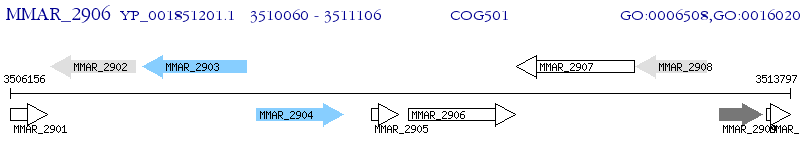
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2906 | - | - | 100% (348) | hypothetical protein MMAR_2906 |
| M. marinum M | MMAR_2614 | - | 3e-07 | 29.91% (107) | hypothetical protein MMAR_2614 |
| M. marinum M | MMAR_5059 | - | 2e-05 | 37.78% (90) | peptidase M48 like- protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv1977 | - | 1e-168 | 83.33% (348) | hypothetical protein Rv1977 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2955 | - | 1e-113 | 66.21% (293) | hypothetical protein MAB_2955 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3476 | - | 1e-167 | 80.80% (349) | peptidase, M48 family protein |
| M. thermoresistible (build 8) | TH_1766 | htpX | 2e-05 | 25.94% (212) | PROBABLE PROTEASE TRANSMEMBRANE PROTEIN HEAT SHOCK PROTEIN |
| M. ulcerans Agy99 | MUL_3147 | - | 1e-07 | 29.91% (107) | hypothetical protein MUL_3147 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2906|M.marinum_M MSQTPATT---RTAFGGISSRAWEHPADRTALSALRRLKGFDQILKLLSG
Rv1977|M.tuberculosis_H37Rv MSQTPATT---RKTFPEISSRAWEHPADRTALSALRRLKGFDQILKLMSG
MSMEG_3476|M.smegmatis_MC2_155 MTRPPATQPPQRTTFPGISSRAWEHPADRTALTALRRLKGFDQVLKVLSG
MAB_2955|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_3147|M.ulcerans_Agy99 MAIVESPP---GHPYPHLPASFYGTPSGSPPGPTGYLEHPRRHPWEIGLL
TH_1766|M.thermoresistible__bu MTWNPHAN-RLKTFVLLVGMSALIVFIGALFQNRSILFLAVLFAVGMNVY
MMAR_2906|M.marinum_M MLRERQHRLLYLASAARVGPRQFADLDALLDECVDVLDAPT--KPEVYVT
Rv1977|M.tuberculosis_H37Rv MLRERQHRLLYLASAARVGPRQFADLDALLDECVDVLDASA--KPELYVM
MSMEG_3476|M.smegmatis_MC2_155 MLRERQHRLLYLASSARVGPRQFADLDELLDDCARVLDAPS--RPEMFVT
MAB_2955|M.abscessus_ATCC_1997 ---------MYLATAIRADDRQFSTLNNTLHDCARILDAPE--IPELYVI
MUL_3147|M.ulcerans_Agy99 VLVIVSSVLLYLVAAAVVLSGKSSLLWISIVATPIVLFALRGLTYGKQRA
TH_1766|M.thermoresistible__bu VYFNSDKLALRAMHAQPVTELQAPVMYRIVRELANTAHQPM---PRLYIS
: : . : . : :
MMAR_2906|M.marinum_M QSPMVNAYTIGMD--APFIVITSGLYDLMTHDELRFVVGHELGHALSGHA
Rv1977|M.tuberculosis_H37Rv QSPIADAFTIGMG--KPFTVITSGLYDLVTHDEMRFVMGHELGHALSGHA
MSMEG_3476|M.smegmatis_MC2_155 QSPVANAYTIGMD--EPFIVITSGMYDLMSHDEMRFVIGHELGHALSGHA
MAB_2955|M.abscessus_ATCC_1997 QGPYANAYTIGMD--RPFIVLTSGLLDLMNEQELRFIVGHELGHALSGHA
MUL_3147|M.ulcerans_Agy99 NGVKMSPTQFAEG--YWLVVEAARRFGLSEVPDAYVVLGNGLINAFSSGH
TH_1766|M.thermoresistible__bu DTANPNAFATGRNPRNAAVCCTTGLLQILNERELRAVLGHELSHVYN---
: .. . . :: : : ::*: * :. .
MMAR_2906|M.marinum_M TYRTMLMHLMRLAGSLGFLPIGGWALRAIVAALLEWQRKSELSGDRAGLL
Rv1977|M.tuberculosis_H37Rv VYRTMMMHLLRLARSFGVLPVGGWALRAIVAALLEWQRKSELSGDRAGLL
MSMEG_3476|M.smegmatis_MC2_155 VYRTMLMHLMRIASTFGFVPIGGWALRAIVAALMEWERKSELSGDRAGLL
MAB_2955|M.abscessus_ATCC_1997 VYRTMLLHLMRLAGNLGWMPIGGWALRAIVAALMEWQRKSELSGDRAGML
MUL_3147|M.ulcerans_Agy99 GFRRYVVVYS------DLFEIRGQARDPEALAFIIGHEVGHLAAGHASYW
TH_1766|M.thermoresistible__bu --RDILISCVAG------------ALAAIVTALANIAMYASMFGGNR--Q
* :: * . . *: . : ...
MMAR_2906|M.marinum_M CCQDLDTAIRVEMKLAGGSRLDKLDSEAF---LAQAAEYERSGDMRDGLL
Rv1977|M.tuberculosis_H37Rv CAQDLDTALRVEMKLAGGCRLDKLDSEAF---LAQAREYETSGDMRDGVL
MSMEG_3476|M.smegmatis_MC2_155 CGQDLDTAIRVELKLAAGARLDKLDSQAF---LAQAREYERAGDMRDGLL
MAB_2955|M.abscessus_ATCC_1997 CGQDVDTAIRVEMKLAGGSRLDEMDTQRF---LAQAAEYERTGDMRDGVL
MUL_3147|M.ulcerans_Agy99 R-QLMQLAMRVPLLGPTLSRSMEYTADNYGYAFRPAGARKAIGVLSAGKY
TH_1766|M.thermoresistible__bu GGPSPFALLLVSFLGPIAATVIRMAVSRS-----REYQADQSGAELTGDP
: * : . . . . . * *
MMAR_2906|M.marinum_M KLLNLELQTHPFSVLRVAELTKWVDAGGYGKVMSGSYPLRLDDGQSSIAD
Rv1977|M.tuberculosis_H37Rv KLLNLELQTHPFSVLRAAALTHWVDTGGYAKVIAGEYPRRADDGNAKFAD
MSMEG_3476|M.smegmatis_MC2_155 KLLNLELKTHPFSVLRAAALTQWVDTGGYGAVMAGDYPRREDDDKAALRD
MAB_2955|M.abscessus_ATCC_1997 KLLNLELQSHPFSVIRAAELSKWIDRGEYGAILSGDYPLRTDDEQTDLGS
MUL_3147|M.ulcerans_Agy99 LLRSVDFDGMADRAGSETGFFVWLAN------MVSSHPVN-TWRAAALRN
TH_1766|M.thermoresistible__bu LALASALRKISGGVQAAP---------------LPPEPQLADQAHLMIAS
: . . . * : .
MMAR_2906|M.marinum_M DLGEAARHYKDGFDQSDDPLIKGIR---DGLGGIVDGVGRAATSAADSLG
Rv1977|M.tuberculosis_H37Rv DLGAAARYYRDGFDQSNDPLIKGIR---DGFGGIVEGVGRAASNAADSLG
MSMEG_3476|M.smegmatis_MC2_155 DIGEAARHYRDGFDQSDDPLIRGIR---DGLGGIVDGVGQAATSAADSVG
MAB_2955|M.abscessus_ATCC_1997 EFRSAARSYKENFDSSTAPLISALRNFGSTLDGVVNVVGQGVTDVATDVR
MUL_3147|M.ulcerans_Agy99 RMVAGSLFFRLKQPALVPPTAWAVP----GDGGWAR--------------
TH_1766|M.thermoresistible__bu PFRAGEKIG--KLFSTHPPIEDRIAR-LEAMAGRGPGHYY----------
: . * : *
MMAR_2906|M.marinum_M RKINEWRHNPK----------------------
Rv1977|M.tuberculosis_H37Rv RKITEWRQPSK----------------------
MSMEG_3476|M.smegmatis_MC2_155 RKISEWRRSAQRDARRDDRRDGPEPDSGSDDAG
MAB_2955|M.abscessus_ATCC_1997 RRFSEWRNNSDDEPDDDT---------------
MUL_3147|M.ulcerans_Agy99 ---------------------------------
TH_1766|M.thermoresistible__bu ---------------------------------