For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVGLVHQDPGIGLTAGEIFAGYTILRLLGSGGMGEVYLAKHPRLPRHEALKVLGAGVCADDDYRRRFIRE ADVAAALWHPNIVRVHDRGEHAGALWISMDYVEGTDAASLLRDHYPVGMPADQVSTIIAAIASALDYAHQ HHDLLHRDVSPANILLSDPGEDGQRILLGDFGIARNIGDASGLTATNMTIGTFPYAAPEQLTDEPIDGRA DQYALAATIYHLLTGTTLFPHTNPAVVISRHLSTPPPALAQTRPELAAFDPVLAVALAKNPADRYARCID FAHSFARAARCGRHPKSSASTMPAPLASRPAKATPAPRTLADRETPQRRGRWRIFAAASTVLALTAVGAS GYSISNDTTASAHRTLAASPEQGPPPSPTQPVAQHPPPPRPMPPVPAPHRVAPPPQRPAETQIISAPPVS RIPAPVVAPPAPPHPPSPDQSFLSLVSEIPGITVTDPATAAATGRAVCTSLQNGGTPHDAAAATVNNTGI TPAQASAGINAAITAYCPQYQG
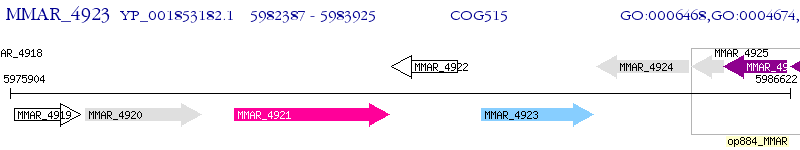
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4923 | pknF_2 | - | 100% (512) | serine/threonine-protein kinase PknF_2 |
| M. marinum M | MMAR_2606 | pknF_3 | e-105 | 55.31% (367) | anchored-membrane serine/threonine-protein kinase PknF_3 |
| M. marinum M | MMAR_2408 | pknF_1 | e-100 | 52.37% (380) | anchored-membrane serine/threonine-protein kinase PknF_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1775 | pknF | 1e-104 | 54.16% (373) | anchored-membrane serine/threonine-protein kinase PKNF |
| M. gilvum PYR-GCK | Mflv_3427 | - | 1e-106 | 51.82% (411) | protein kinase |
| M. tuberculosis H37Rv | Rv1746 | pknF | 1e-103 | 52.34% (384) | anchored-membrane serine/threonine-protein kinase PKNF |
| M. leprae Br4923 | MLBr_00016 | pknB | 7e-45 | 36.26% (342) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_4435 | - | 3e-62 | 41.71% (350) | serine/threonine-protein kinase PknJ |
| M. avium 104 | MAV_3145 | - | 2e-99 | 51.83% (382) | PknF protein |
| M. smegmatis MC2 155 | MSMEG_3677 | - | 1e-108 | 48.37% (490) | serine/threonine protein kinase |
| M. thermoresistible (build 8) | TH_1607 | pknI | 5e-78 | 65.78% (225) | PROBABLE TRANSMEMBRANE SERINE/THREONINE-PROTEIN KINASE I |
| M. ulcerans Agy99 | MUL_0482 | pknF_2 | 0.0 | 99.00% (500) | serine/threonine-protein kinase PknF_2 |
| M. vanbaalenii PYR-1 | Mvan_5635 | - | 1e-105 | 58.70% (339) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4923|M.marinum_M --------------------------------------------------
MUL_0482|M.ulcerans_Agy99 --------------------------------------------------
Mb1775|M.bovis_AF2122/97 --------------------------------------------------
Rv1746|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_3677|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3427|M.gilvum_PYR-GCK --------------------------------------------------
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_3145|M.avium_104 MNEHKGTTRRIGAGRRLLSNPRQARAALRAGFQSLPGMPIGQLFRRIPPV
MAB_4435|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00016|M.leprae_Br4923 --------------------------------------------------
MMAR_4923|M.marinum_M -----------MVGLVHQDPGIGLTAGEIFAGYTILRLLGSGGMGEVYLA
MUL_0482|M.ulcerans_Agy99 -----------------------MTAGEIFAGYTILRLLGSGGMGEVYLA
Mb1775|M.bovis_AF2122/97 ---------------------MPLAEGSTFAGFTIVRQLGSGGMGEVYLA
Rv1746|M.tuberculosis_H37Rv ---------------------MPLAEGSTFAGFTIVRQLGSGGMGEVYLA
MSMEG_3677|M.smegmatis_MC2_155 ---------------------MPLAAGETFAGYNIIRLLGSGGMGEVYLA
Mflv_3427|M.gilvum_PYR-GCK ---------------------MPLAAGETFAGYTIVRLLGSGGMGEVYLA
TH_1607|M.thermoresistible__bu ---------------------MPLADGQQFAEYTILRRLGAGGMGEVYLA
Mvan_5635|M.vanbaalenii_PYR-1 ---------------------MPLSDGEVIAGYTIVRMLGSGGMGKVYLA
MAV_3145|M.avium_104 DSRPHPEPAAAQGNSEPGGPRLTIGNGASFAGYTILRQLGAGGMAEVYLA
MAB_4435|M.abscessus_ATCC_1997 -----MTSSDRSSPGPGGQQPGDLPPGTVVSGYQIERVLGRGGMGTVYLA
MLBr_00016|M.leprae_Br4923 ----------------------MTTPPHLSDRYELGDILGFGGMSEVHLA
: : ** ***. *:**
MMAR_4923|M.marinum_M KHPRLPRHEALKVLGAGVCADDDYRRRFIREADVAAALWHPNIVRVHDRG
MUL_0482|M.ulcerans_Agy99 KHPRLPRHEALKVLGAGVCADDDYRRRFIREADVAAALWHPNIVRVNDRG
Mb1775|M.bovis_AF2122/97 RHPRLPRQDALKVLRADVSADGEYRARFNREADAAASLWHPHIVAVHDRG
Rv1746|M.tuberculosis_H37Rv RHPRLPRQDALKVLRADVSADGEYRARFNREADAAASLWHPHIVAVHDRG
MSMEG_3677|M.smegmatis_MC2_155 AHPRLPRQDALKVLPTSISADPEYRERFNREADVAAALWHPHIVEIHDRG
Mflv_3427|M.gilvum_PYR-GCK QHPRLPRRDALKVLPAAVSADVEYRKRFEREADIAATLWHPHIVGVHDRG
TH_1607|M.thermoresistible__bu RHPRLPRNDALKVLSAEVSADDEYRRRFEQEADIAATLWHPHIVAVHDRG
Mvan_5635|M.vanbaalenii_PYR-1 KHPRLPRYDALKVLSTTVCADSEYRERFHREADIAATLWHPHIVAVHDRG
MAV_3145|M.avium_104 LHPRLPRRDVIKVLAEAVTVDPEFRERFNREADLAATLWHPHIVGVHDRG
MAB_4435|M.abscessus_ATCC_1997 ANPNLQRSEALKILSAQSSRDPEFRARFMREASLAASLDHPNIVTVHNRG
MLBr_00016|M.leprae_Br4923 RDIRLHRDVAVKVLRADLARDPSFYLRFRREAQNAAALNHPSIVAVYDTG
. .* * .:*:* * .: ** :**. **:* ** ** : : *
MMAR_4923|M.marinum_M EH----AGALWISMDYVEGTDAASLLRDHYPVGMPADQVSTIIAAIASAL
MUL_0482|M.ulcerans_Agy99 EH----AGALWISMDDVEGTDAASLLRDHYPVGMPADQVSTIIAAIANAL
Mb1775|M.bovis_AF2122/97 EF----DGQLWIDMDFVDGTDTVSLLRDRYPNGMPGPEVTEIITAVAEAL
Rv1746|M.tuberculosis_H37Rv EF----DGQLWIDMDFVDGTDTVSLLRDRYPNGMPGPEVTEIITAVAEAL
MSMEG_3677|M.smegmatis_MC2_155 EH----DGQLWIAMDYVDGTDAARLLKEQYSGGMPPHQVCEIVTAMAGAL
Mflv_3427|M.gilvum_PYR-GCK ED----AGRIWISMDYVEGADAAHLLAEDYRTGLPVAEVAQIVTAVADAL
TH_1607|M.thermoresistible__bu EY----DGQLWIAMDHVDGTDVGRLLGEQYPNGMPPDAVIRIITAVADAL
Mvan_5635|M.vanbaalenii_PYR-1 EY----EGRLWISMDHVEGTDAAWLLAERYPHGMPPALVIRIVTAVGEAL
MAV_3145|M.avium_104 EF----NGHLWISMDYVEGTDASRLVKESYADGMPLDEVSAIVQAVAGAL
MAB_4435|M.abscessus_ATCC_1997 ETP---EGDLWIAMQYVQGSDAH---SESASGRMSLTRAVHIISEVAKAL
MLBr_00016|M.leprae_Br4923 EAETSAGPLPYIVMEYVDGATLRDIVHTDGP--MPPQQAIEIVADACQAL
* :* *: *:*: :. . *: **
MMAR_4923|M.marinum_M DYAHQHHDLLHRDVSPANILLSDPGEDG--QRILLGDFGIARNIGDASGL
MUL_0482|M.ulcerans_Agy99 DYAHQHHDLLHRDVSPANILLSDPGEDG--QRILLGDFGIARNIGDASGL
Mb1775|M.bovis_AF2122/97 DYAHERR-LLHRDVKPANILIANPDSPD--RRIMLADFGIAGWVDDPSGL
Rv1746|M.tuberculosis_H37Rv DYAHERR-LLHRDVKPANILIANPDSPD--RRIMLADFGIAGWVDDPSGL
MSMEG_3677|M.smegmatis_MC2_155 DYAHQRG-LLHRDVKPANILLGHPGTDD--QRIMLADFGIARYADEASGL
Mflv_3427|M.gilvum_PYR-GCK DYAHDRH-LLHRDVKPANILIARPDSNT--RRIMLADFGIARWDNDISGL
TH_1607|M.thermoresistible__bu DHAHSRN-LLHRDVKPANILLAAGDGEQGAERVLLADFGIARWIDRSSGL
Mvan_5635|M.vanbaalenii_PYR-1 DYAHQRG-LLHRDVKPANILIADPETEN--ERIMLADFGIARRVGEVSTL
MAV_3145|M.avium_104 DYAHARG-LLHRDVKPANILLTHPEAGE--RRILLADFGVARHLGDISGI
MAB_4435|M.abscessus_ATCC_1997 DYLHARG-LVHRDVKPANILLS---AGD--RRIMLGDFGLTRALDDSNSV
MLBr_00016|M.leprae_Br4923 NFSHQNG-IIHRDVKPANIMISATNAVK------VMDFGIARAIADSTSV
:. * . ::****.****:: : ***:: . :
MMAR_4923|M.marinum_M TATNMTIGTFPYAAPEQLTDEP-IDGRADQYALAATIYHLLTGTTLFPHT
MUL_0482|M.ulcerans_Agy99 TATNMTIGTFPYAAPEQLTDEP-IDGRADQYALAATVYHLLTGTTLFPHT
Mb1775|M.bovis_AF2122/97 TATNMTVGTVSYAAPEQLMGNE-LDGRADQYALAATAFHLLTGSPPFQHA
Rv1746|M.tuberculosis_H37Rv TATNMTVGTVSYAAPEQLMGNE-LDGRADQYALAATAFHLLTGSPPFQHA
MSMEG_3677|M.smegmatis_MC2_155 TATNMTVGTVTYAAPEQLLGNH-LDGRADQYALAATAYQLLTGRTPFEHS
Mflv_3427|M.gilvum_PYR-GCK TATNMTVGTVSYAAPEQLMGQD-LDGRADQYALAATAFHLLTGSPPFSHS
TH_1607|M.thermoresistible__bu TETDMTVGTVAYAAPEQLRAAD-IDGRADQYALAATAFQLMTGAPPFQHS
Mvan_5635|M.vanbaalenii_PYR-1 TGTSMTVGTVAYSAPEQLTADEHIDGRADQYALAATAFQLLTGSPPFQHS
MAV_3145|M.avium_104 TETNVAVGTVAYAAPEQLTGSP-IDGRADQYALAATAFHLLTGAPPFQHS
MAB_4435|M.abscessus_ATCC_1997 TSAGNVTATIAYAAPEILSGTA-VDGRADVYALGCTLFRMLTGQPPFGSG
MLBr_00016|M.leprae_Br4923 TQTAAVIGTAQYLSPEQARGDS-VDARSDVYSLGCVLYEILTGEPPFIGD
* : . .* * :** :*.*:* *:*... :.::** . *
MMAR_4923|M.marinum_M N-PAVVISRHLSTPPPALAQTRP-ELAAFDPVLAVALAKNPADRYARCID
MUL_0482|M.ulcerans_Agy99 N-PAVVISRHLSTPPPALAQTRP-ELAAFDPVLAVALAKNPADRYARCID
Mb1775|M.bovis_AF2122/97 N-PAVVISQHLSASPPAIGDRVP-ELTPLDPVFAKALAKQPKDRYQRCVD
Rv1746|M.tuberculosis_H37Rv N-PAVVISQHLSASPPAIGDRVP-ELTPLDPVFAKALAKQPKDRYQRCVD
MSMEG_3677|M.smegmatis_MC2_155 N-PAVVISQHLSAAPPAVGTHRP-DLATLDPVFAKALAKDPRDRYARCAD
Mflv_3427|M.gilvum_PYR-GCK N-PAVVISRHLNSAPATVAAHRP-ELAAVDPVLTRALAKNPADRYPRCAD
TH_1607|M.thermoresistible__bu N-PAXXXCGR----------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 N-PAIVISQHLTAQPPSISVHRP-ELSSLGMAFEKALAKSPADRFDRCVD
MAV_3145|M.avium_104 N-PIAVIGQHLHEDPPRLSDFRP-ELAALDEVFCQALAKVPEDRFDRCRA
MAB_4435|M.abscessus_ATCC_1997 NSLPAIINAHLSAPPPRISQLAP-VPESWDALIAKAMAKTPADRYQSAGE
MLBr_00016|M.leprae_Br4923 S-PVSVAYQHVREDPIPPSQRHEGISVDLDAVVLKALAKNPENRYQTAAE
. :
MMAR_4923|M.marinum_M FAHSFARA------ARCGRHPKS---SASTMPAPLASRPAKATPAPRTLA
MUL_0482|M.ulcerans_Agy99 FAHVFARA------ARCGRHPKS---SASTMPAPLASRPAKATPAPRTLA
Mb1775|M.bovis_AF2122/97 FARALG--------HRLG------------GAGDPDDTRVSQPVAVAAPA
Rv1746|M.tuberculosis_H37Rv FARALG--------HRLG------------GAGDPDDTRVSQPVAVAAPA
MSMEG_3677|M.smegmatis_MC2_155 FARALR--------HRLGTDA---------DVADPDATRLAPVARHAAPH
Mflv_3427|M.gilvum_PYR-GCK FAREFN--------ARLD---------------------------AGAPA
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 FARALA--------NRSGTELPG-------SAFDPDATRHAAAPTGPRHA
MAV_3145|M.avium_104 FAAAVS--------RECDGAAAIGPDARSRTVASPAHRRRGPGRVIAAVT
MAB_4435|M.abscessus_ATCC_1997 LAMAAQQILTD--PAQHRQTVTQPVPATTPAPIAAPEPVHAMAPLAMADT
MLBr_00016|M.leprae_Br4923 MRADLIRVHSGQPPEAPKVLTDADRSCLLSSGAGNFGVPRTDALSRQSLD
MMAR_4923|M.marinum_M DRETPQRRGRWRIFAAASTVLALTAVGASGYSISNDTTASAHRTLAASPE
MUL_0482|M.ulcerans_Agy99 DRETPQRRGRWRIFAAASTVLALTAVGASGYSISNDTTASAHRTLAASPE
Mb1775|M.bovis_AF2122/97 KRSLLRTAVIVPAVLAMLLVMAVAVTVREFQRADDERAAQPARTRTTTSA
Rv1746|M.tuberculosis_H37Rv KRSLLRTAVIVPAVLAMLLVMAVAVAVREFQRADDERAAQPARTRTTTSA
MSMEG_3677|M.smegmatis_MC2_155 KR--PKTAALIAGVLGVLLVAAIVVAAVEFRRADDERPVARASGAATTTS
Mflv_3427|M.gilvum_PYR-GCK HTSRIRPAVLIPVVLAVLLVAAVAVAAFEFLRADR---ADRADRADPPPM
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 KTAAAPRSRRRVVLAGAGLVLALIAVAAAVFLARFERDVGRGEARATASA
MAV_3145|M.avium_104 HRFSPQTRWAAALVCAVLVAVAATWSVLYSFQPGAPPANPALASKPSPPA
MAB_4435|M.abscessus_ATCC_1997 GQSMLKHKRIAAIIGVMVLVVASSVVAVLIWPRADEGTDPRTKASPSAAQ
MLBr_00016|M.leprae_Br4923 ETESDGSIGRWVAVVAVLAVLTIAIVAAFNTFGGNTRDVQVPDVRGQVSA
MMAR_4923|M.marinum_M QGPPPSPTQPVAQHPPPPRP-----------------------MPPVPAP
MUL_0482|M.ulcerans_Agy99 QGPPPSPTQPVAQHPPPPRP-----------------------MPPVPAP
Mb1775|M.bovis_AF2122/97 GTTTSVAPASTTRP-----------------------------APTTPTT
Rv1746|M.tuberculosis_H37Rv GTTTSVAPASTTRP-----------------------------APTTPTT
MSMEG_3677|M.smegmatis_MC2_155 HTTTPVTTAPASPPPEVTDTVT---------------------ITDTATD
Mflv_3427|M.gilvum_PYR-GCK ARTTTITTS-----------------------------------PTTSTT
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 TAST------------------------------------------APTG
MAV_3145|M.avium_104 AVAAPIAGGPVLNGTYKLDYDQTKRTTNGIGIRHDGAGTNWWAFRSACTS
MAB_4435|M.abscessus_ATCC_1997 VAPGASLAGQPNTVLDTLLPGQFDYPEG---------------WEKNPSP
MLBr_00016|M.leprae_Br4923 DAISALQNRGFKTRTLQKPDSTIPPDHVISTEPGANASVG---AGDEITI
MMAR_4923|M.marinum_M HRVAPPPQ----------------RPAETQIIS-----------APPVSR
MUL_0482|M.ulcerans_Agy99 HRVAPPPQ----------------RPAETQIIS-----------APPVSR
Mb1775|M.bovis_AF2122/97 TGAADTAT--------------ASPTAAVVAIG-----------ALCFPL
Rv1746|M.tuberculosis_H37Rv TGAADTAT--------------ASPTAAVVAIG-----------ALCFPL
MSMEG_3677|M.smegmatis_MC2_155 TAAVDTSTPATTTAATTA----AAAAAQTAVIG-----------ASCSPV
Mflv_3427|M.gilvum_PYR-GCK TATP-------------------PATPASVVIG-----------ANCAPL
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 SGSV---------------------TLPVVVVG-----------ADCATL
MAV_3145|M.avium_104 SGCAATGTRLDDATHQAAGGPDGDQTDTLRFVGGYWQGAPEQQRVGCTRP
MAB_4435|M.abscessus_ATCC_1997 TFRSGEFN-------------PAVPPGSATNIGG--------TPVGCNTI
MLBr_00016|M.leprae_Br4923 NVSTGPEQREVPDVSSLNYTDAVKKLTSSGFKSFKQANSPSTPELLGKVI
MMAR_4923|M.marinum_M IPAPVVAP-PAPPHPPSPDQSFLSLVSEIPGITVT---------------
MUL_0482|M.ulcerans_Agy99 IPAPVVAP-PAPPHPPSPDQSFLSLVSEIPGITVT---------------
Mb1775|M.bovis_AF2122/97 GSTGTTKT-GATAYCSTLQGTNTTIWSLTEDTVAS---------------
Rv1746|M.tuberculosis_H37Rv GSTGTTKT-GATAYCSTLQGTNTTIWSLTEDTVAS---------------
MSMEG_3677|M.smegmatis_MC2_155 GATATTSD-GSTAYCSTLATTGASIWSLTQGTIPS---------------
Mflv_3427|M.gilvum_PYR-GCK DATATTGD-GTTAYCAAVPGSAGTIWSLTQGTLPA---------------
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 GAAGVTEA-GDPAYCARLASSGAPLWSLYPGEIPH---------------
MAV_3145|M.avium_104 GGPAGATQ-QETIARSLAPQSDGTLRGTETETVLSNECGAQGAVVRVPVV
MAB_4435|M.abscessus_ATCC_1997 DPAWALRAKGRETSTLYLRDRKKPEREILIRVVPAADALSTDGASDALKR
MLBr_00016|M.leprae_Br4923 GTNPSANQTSAITNVITIIVGSGPETKQIPDVTGQIVEIAQKNLNVYGFT
MMAR_4923|M.marinum_M ----------DPATAAATGRAVCTSLQNGGTPHD------AAAATVNNTG
MUL_0482|M.ulcerans_Agy99 ----------DPATAAATGRAVCTSLQNGGTPHD------AAAATVNNTG
Mb1775|M.bovis_AF2122/97 -----------PTVTATADPTEAPLPIEQESPIR---------VCMQQTG
Rv1746|M.tuberculosis_H37Rv -----------PTVTATADPTEAPLPIEQESPIR---------VCMQQTG
MSMEG_3677|M.smegmatis_MC2_155 -----------PTVTPAA--TDEPLPFAEESPVR---------VCMQQTG
Mflv_3427|M.gilvum_PYR-GCK -----------PTSTPPPDPTEQPLSDAEAFPVL---------TCMEQTG
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 -----------PTGTSAP----EGESQGPDTPVL---------VCMQQTG
MAV_3145|M.avium_104 ATRVGDVPPGVTVADPASVINASPTATAPAPPVLGGLCSDVGKVAYDPTN
MAB_4435|M.abscessus_ATCC_1997 CDAVAYIIPGSSTTWRCSFEFDEPNPVQNGQKELT----TTESCYMFPSG
MLBr_00016|M.leprae_Br4923 KFSQASVDSPRPTGEVIGTNPPKDATVPVDSVIELQVSKGNQFVMPDLSG
MMAR_4923|M.marinum_M ITPAQAS-------------AGINAAITAYCPQYQG--------------
MUL_0482|M.ulcerans_Agy99 ITPAQAS-------------AGINAAITAYCPQYQG--------------
Mb1775|M.bovis_AF2122/97 QTRRECR-------------EEIRRSNGWP--------------------
Rv1746|M.tuberculosis_H37Rv QTRRECR-------------EEIRRSNGWP--------------------
MSMEG_3677|M.smegmatis_MC2_155 MTRRECR-------------EAIRRSNGLP--------------------
Mflv_3427|M.gilvum_PYR-GCK QTRRECR-------------RAIRESNGQPPLP-----------------
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 QSQVDCH-------------DDILQENTDPGANDTQTPG-----------
MAV_3145|M.avium_104 NEQIVCEGSSWAKAPITMGVHAAGSSCDRPGTSVFAMSTSSDGYLLQCDP
MAB_4435|M.abscessus_ATCC_1997 ANSIRQHVSFNVVRGLLVYILASGASNRNDAAGSLSATALSFATTMKILR
MLBr_00016|M.leprae_Br4923 MFWADAEPRLRALGWTGVLDKGPDVDAGGSQHNRVAYQNPPAGAGVNRDG
MMAR_4923|M.marinum_M ----------
MUL_0482|M.ulcerans_Agy99 ----------
Mb1775|M.bovis_AF2122/97 ----------
Rv1746|M.tuberculosis_H37Rv ----------
MSMEG_3677|M.smegmatis_MC2_155 ----------
Mflv_3427|M.gilvum_PYR-GCK ----------
TH_1607|M.thermoresistible__bu ----------
Mvan_5635|M.vanbaalenii_PYR-1 ----------
MAV_3145|M.avium_104 VTRTWTRPAG
MAB_4435|M.abscessus_ATCC_1997 YGR-------
MLBr_00016|M.leprae_Br4923 IITLKFGQ--