For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTKRVAVNRQPSRAELERLLSADLRAITAHSDRVGRYYARHNDLSHGDFQALLHIMVAETAGTPVTPAQL SQRLDVSAAAITYLADRMIHAGHIRREADPDDRRKSHLRFEQSSLELAHKFFMPLGDHMREAMADLSDRD LRAAHRVLTAMMDGMETFEKELYESPAPTSGRPRKTPAQAESP
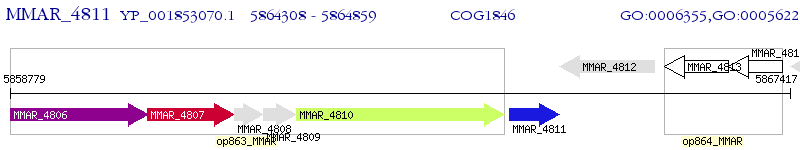
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4811 | - | - | 100% (183) | MarR family transcriptional regulator |
| M. marinum M | MMAR_1675 | - | 4e-39 | 51.66% (151) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0223 | - | 2e-35 | 48.00% (150) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3893 | - | 2e-58 | 62.98% (181) | transcriptional regulator, MarR family protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3314 | - | 3e-06 | 25.38% (130) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0379 | - | 1e-100 | 98.36% (183) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0684 | - | 4e-41 | 50.62% (160) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4811|M.marinum_M MTKRVAVNRQPSRAELERLLSADLRAITAHSDRVGRYYARHNDLSHGDFQ
MUL_0379|M.ulcerans_Agy99 MTKRVAVNRQPSRAELERLLSADLRSITAHSDRVGRYYARHNDLSHGDFQ
MAV_3893|M.avium_104 MTKGGAADRRLDRTELEKLMSADMRAITAQSDRIGRHFARQNNVSGTDFH
Mflv_0223|M.gilvum_PYR-GCK MQN-------GDRAQVEALIAADVRALSAASDAIGQSFAALHGLTASDFR
Mvan_0684|M.vanbaalenii_PYR-1 MQD-------VDRVALEASIAVDVRALTAESDRIGHRFAGLHALTPNDFR
TH_3314|M.thermoresistible__bu VTT-------SDRPQILAELNAEFRVQALRSVMLHSAVAARLGIAVTDFN
: .* : : .:.* : * : * :: **.
MMAR_4811|M.marinum_M ALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIHAGHIRREAD
MUL_0379|M.ulcerans_Agy99 ALLHIMVAETAGTPVTPAQLSQRLDVS-AAAITYLADRMIHAGHIRREAD
MAV_3893|M.avium_104 ALLHIMVAETTGTPLTPAQLRQRMDVS-PAAITYLVDRMIDAGHIRREPD
Mflv_0223|M.gilvum_PYR-GCK ALLHVTVAEDDGAPLTAGELRARMGTS-GAAITYLVERMIESGHVLRDTD
Mvan_0684|M.vanbaalenii_PYR-1 ALLHVMVAENAGTPLTAGELRNRMGTS-GAAITYLVERMNASGHLRRESD
TH_3314|M.thermoresistible__bu CLNVLSMEG----PLTAGQLAERTDLTRGGAVTAMIDRLEAAGFVHRRRD
.* : : *:*..:* * . : .*:* : :*: :*.: * *
MMAR_4811|M.marinum_M PDDRRKSHLRFEQSSLELAHKFFMPLGDHMREAMADLSDRDLRAAHRVLT
MUL_0379|M.ulcerans_Agy99 PDDRRKSHLRFEQSSLELAHRFFMPLGDHMREAMADLSDRDLRAAHRVLT
MAV_3893|M.avium_104 PQDRRKTLLRYEKPGMALAHSFFTPLGAELRDALAHLSDRDLAAAHRVFT
Mflv_0223|M.gilvum_PYR-GCK PTDRRKVMLRVADHGMAVARDFFSPLSAHTHAALADLPDEDLLAAHRVFA
Mvan_0684|M.vanbaalenii_PYR-1 PRDRRKVMLRVADHGMDVAREFFTPLSDHVHEGLAGLPDGDLAAAHRVFT
TH_3314|M.thermoresistible__bu DDDRRRVLVELDEAAAARIAPLFAGLGESLNDHLDSYRTDELRLLLDAVR
***: :. . . :* *. . : :* ..
MMAR_4811|M.marinum_M AMMDGMETFEKELYES---PAPTSGRPRKTPAQAESP--
MUL_0379|M.ulcerans_Agy99 AMMYGMETFEKELYES---PAPTSGRPRKTPAQAESP--
MAV_3893|M.avium_104 AMIEAMTAFESRLAASTSKPPAAPGRKRATTAGRRGAPR
Mflv_0223|M.gilvum_PYR-GCK ALTAAMVSFGAELG---------SGRLKTDD--------
Mvan_0684|M.vanbaalenii_PYR-1 ALIDAMSTFRAALGAD---SGGSSGRLENR---------
TH_3314|M.thermoresistible__bu GINQRVARATDALREG-----------------------
.: : *