For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRFSYAEAMTDPSFYVPLAQAAEAAGYHSMTIADSLAYPYQSDSKYPYTPDGNREFLDGKEIIETFVLTA ALGAVTKTLRFNFFVLKLPVRPPALVAKQAGSLAALIGNRVGLGVGTSPWPEDYELMGVPFAKRGKRIDE CIEIVKGLTTGEYFEFHGEFYDIPKTKMSPAPTAPIPILVGGHADAALRRAARLDGWMHGGGTDPDELDR LITKLQHFREEEGHTGPFEIHVISMDAYTVDGIKRLQDKGVTDVIVGFRMPYIKGPDTEPLQTKIAHLEK FAEKIISKV
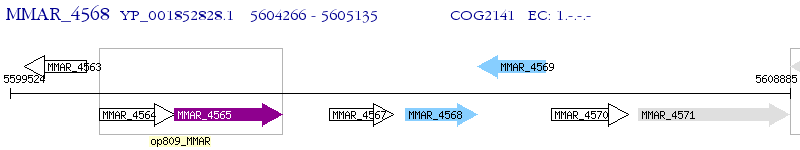
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4568 | - | - | 100% (289) | oxidoreductase |
| M. marinum M | MMAR_5009 | - | 6e-54 | 40.89% (291) | oxidoreductase |
| M. marinum M | MMAR_3291 | - | 4e-39 | 36.97% (284) | hypothetical protein MMAR_3291 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0965c | - | 1e-145 | 84.78% (289) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1850 | - | 1e-133 | 76.21% (290) | hypothetical protein Mflv_1850 |
| M. tuberculosis H37Rv | Rv0940c | - | 1e-145 | 84.78% (289) | oxidoreductase |
| M. leprae Br4923 | MLBr_00269 | - | 3e-08 | 25.00% (200) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2449 | - | 1e-137 | 80.34% (290) | hypothetical protein MAB_2449 |
| M. avium 104 | MAV_1061 | - | 1e-143 | 82.70% (289) | hypothetical protein MAV_1061 |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 3e-27 | 31.40% (258) | hydride transferase 1 |
| M. thermoresistible (build 8) | TH_2326 | - | 1e-140 | 80.62% (289) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4083 | - | 3e-54 | 40.89% (291) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4885 | - | 1e-134 | 78.35% (291) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0965c|M.bovis_AF2122/97 -----MRFSYAEAMTDFTFYIPLAKAAEAAGYSSMTIPDSIAYPFESDSK
Rv0940c|M.tuberculosis_H37Rv -----MRFSYAEAMTDFTFYIPLAKAAEAAGYSSMTIPDSIAYPFESDSK
MMAR_4568|M.marinum_M -----MRFSYAEAMTDPSFYVPLAQAAEAAGYHSMTIADSLAYPYQSDSK
Mflv_1850|M.gilvum_PYR-GCK -----MRFTFAEAMTDPTYYIPLAKAAEAAGYHAMTIPDSIAYPFESDSK
Mvan_4885|M.vanbaalenii_PYR-1 -----MRFTYAEAMTDPTFYIPLAQAAEAAGYHAMTIADSVAYPFESDSK
TH_2326|M.thermoresistible__bu -----MRFTYAEAMTDPSYYIPLAQAAEAAGYHAMTIPDSVAYPFESDSK
MAV_1061|M.avium_104 -----MRFTYAEAMTDFRYYIPLAKAAEAAGYHAMTIADSIAYPFESDSK
MAB_2449|M.abscessus_ATCC_1997 -----MRFTFAEAMTDPSYYIPLAQAAEAAGYDGMTIADSLAYPYESDAK
MUL_4083|M.ulcerans_Agy99 -----MKYTVSIAMGPIDELIGIAKTAEEVGFDSIALPDSLFYMEKAAAD
MSMEG_2811|M.smegmatis_MC2_155 -MQISLHALGIGAGARREIIDAVAVAAERAGFTTLWCGEHVVMVDRPRSR
MLBr_00269|M.leprae_Br4923 MAELRLGYKASAEQFAPRELVELGVAAEAHGMDSATVSDHFQPWRHQGGH
: :. :** * : . . .
Mb0965c|M.bovis_AF2122/97 YPYTPDGNREFMDGKPFIETFVLTAALGAVTTRLRFNFFVLKLPIRP-PA
Rv0940c|M.tuberculosis_H37Rv YPYTPDGNREFMDGKPFIETFVLTAALGAVTTRLRFNFFVLKLPIRP-PA
MMAR_4568|M.marinum_M YPYTPDGNREFLDGKEIIETFVLTAALGAVTKTLRFNFFVLKLPVRP-PA
Mflv_1850|M.gilvum_PYR-GCK YPYTPDGNREFLDGKAFIESFVLTAALCAVTTKLHFNHFVLKLPIRP-PA
Mvan_4885|M.vanbaalenii_PYR-1 YPYTADGNREFLDGKAFIESFVLTAALCAVTTKLHFNHFVLKLPIRP-PA
TH_2326|M.thermoresistible__bu YPYTPDGDREFLDGKAFIESFVLASALCAATTTLRFNFFVLKLPIRP-PA
MAV_1061|M.avium_104 YPYTPDGSREFLDGKEFIETFVLTSALGAVTSKLRFNFFVLKLPIRP-PA
MAB_2449|M.abscessus_ATCC_1997 YPYTPDGNREFLEGKPFIEALTLTAALGAVTSTLRFNIFVLKLPIRP-PA
MUL_4083|M.ulcerans_Agy99 YPYTPDGSRMWNQDTPWVDPLIAAAAMGAATSKLRFYTNVLKLGSRN-PL
MSMEG_2811|M.smegmatis_MC2_155 YPYSADGRIAVAADADWLDPMIALSFAAAATSTIRIATGVLLLPEHN-PV
MLBr_00269|M.leprae_Br4923 ASFS----------------LSWMTAVGERTNRILLGTSVLTPTFRYNPA
.:: : : *. : : ** : *
Mb0965c|M.bovis_AF2122/97 LVAKQAGSLAALIGNRVGLGVGTSPWP----EDYELMGVPFAKRGKRIDE
Rv0940c|M.tuberculosis_H37Rv LVAKQAGSLAALIGNRVGLGVGTSPWP----EDYELMGVPFAKRGKRIDE
MMAR_4568|M.marinum_M LVAKQAGSLAALIGNRVGLGVGTSPWP----EDYELMGVPFAKRGKRIDE
Mflv_1850|M.gilvum_PYR-GCK LVAKQAGSLAALFDNRLGLGVGTSPWP----EDYELLNVPFARRGKRMDE
Mvan_4885|M.vanbaalenii_PYR-1 LVAKQAGSLAALFDNRLGLGVGTSPWP----EDYELMNVPFAKRGKRMDE
TH_2326|M.thermoresistible__bu LVAKQAGSLAAMFDNRLGLGVGTSPWP----EDYEVMGVPYAKRGKRMDE
MAV_1061|M.avium_104 LVAKQIGSLAAMTDNRVGFGVGTSPWP----EDYELLGVPFARRGKRMDE
MAB_2449|M.abscessus_ATCC_1997 LVAKQASSIAALTGNRLGLGVGTSPWP----EDYELMNVPFAKRGKRMDE
MUL_4083|M.ulcerans_Agy99 LLARQVGSVANLTNNRFGFGVGIGWAP----EEFEWCGVPYRRRGARVDE
MSMEG_2811|M.smegmatis_MC2_155 IMAKQAASVDQLSGGRLLLGVGIGWSR----EEFEALGVPFARRGARTAE
MLBr_00269|M.leprae_Br4923 VIGQAFATMGCLYPNRVFLGVGTGEALNEVATGYQGAWPEFKERFARLRE
::.: .:: : .*. :*** . :: : .* * *
Mb0965c|M.bovis_AF2122/97 CIEIVRGLTTGDYFEFHGEFYDIPKTKMTPAPTQ-PIPILVGGHADAALR
Rv0940c|M.tuberculosis_H37Rv CIEIVRGLTTGDYFEFHGEFYDIPKTKMTPAPTQ-PIPILVGGHADAALR
MMAR_4568|M.marinum_M CIEIVKGLTTGEYFEFHGEFYDIPKTKMSPAPTA-PIPILVGGHADAALR
Mflv_1850|M.gilvum_PYR-GCK CIDIIRGLTSGDYFEYHGEFYDIPKTKMTPAPSK-PVPILIGGHADAALR
Mvan_4885|M.vanbaalenii_PYR-1 CIDIIRGLTSGDYFEYHGEFYDIPKTKMTPAPTK-PIPILIGGHADAALR
TH_2326|M.thermoresistible__bu CIDIIRGLTTGDYFEYHGEFYDIPKTKMTPAPSK-PIPILIGGHADAALR
MAV_1061|M.avium_104 CIEIIQGLTTGEYFEFHGEFYDIPKTKMTPAPTK-PIPVLVGGHADAALR
MAB_2449|M.abscessus_ATCC_1997 CIEIIQGLTTGDYFEFHGEFYDIPKTKMCPAPTA-PIPILVGGHADAALR
MUL_4083|M.ulcerans_Agy99 MIDVIKLVLGGGMVEFHGQFYDFDRLQMSPAPSE-PVPFYVGGHTDVALR
MSMEG_2811|M.smegmatis_MC2_155 YVEAMRTLWRQDVADFTGEFAAFDAVRLHPKPSAGSIPIIVGGNGDRALA
MLBr_00269|M.leprae_Br4923 SVRLMRELWRGDRVDFDGDYYQLKGASIYDVPEG-GVPIYIAAGGPEVAK
: :: : :: *:: : : * :*. :.. .
Mb0965c|M.bovis_AF2122/97 RAARA-DGWMHGGG-----------------------DPDELDRLIARVK
Rv0940c|M.tuberculosis_H37Rv RAARA-DGWMHGGG-----------------------DPDELDRLIARVK
MMAR_4568|M.marinum_M RAARL-DGWMHGGGT----------------------DPDELDRLITKLQ
Mflv_1850|M.gilvum_PYR-GCK RAARN-DGWMHGGG-----------------------DPEELDALLAKLT
Mvan_4885|M.vanbaalenii_PYR-1 RAARN-DGWMHGGG-----------------------DPEELDALLGKLN
TH_2326|M.thermoresistible__bu RAARN-DGWMHGGG-----------------------DGEELDHLLAKLK
MAV_1061|M.avium_104 RAARL-DGWMHGGG-----------------------DPEELDHLLAKLK
MAB_2449|M.abscessus_ATCC_1997 RAARC-DGWMHGGG------------------------TEDLDELLATLN
MUL_4083|M.ulcerans_Agy99 RAARIGDGWTSAMT-----------------------TGEQLAETIGKLN
MSMEG_2811|M.smegmatis_MC2_155 RVASWGDGWYGFNLD----------------------GVAAVGERMDMLR
MLBr_00269|M.leprae_Br4923 YAGRAGEGFVCTSGKGEELYTEKLIPAVLEGAAVAGRDADDIDKMIEIKM
.. :*: : :
Mb0965c|M.bovis_AF2122/97 RLREEAGKTS--PFEIHVISLDGFTVDGVKR-LEDKGVTDVIVGFRVPYT
Rv0940c|M.tuberculosis_H37Rv RLREEAGKTS--PFEIHVISLDGFTVDGVKR-LEDKGVTDVIVGFRVPYT
MMAR_4568|M.marinum_M HFREEEGHTG--PFEIHVISMDAYTVDGIKR-LQDKGVTDVIVGFRMPYI
Mflv_1850|M.gilvum_PYR-GCK RFRDEAGTAGADDYQIHVISLDAYSLDGIKR-LEDKGVTDVIVGFRIPYI
Mvan_4885|M.vanbaalenii_PYR-1 RFREEAGTADNADYQIHVISIDAYTADGIKR-LEDKGVTDVIVGFRIPYI
TH_2326|M.thermoresistible__bu RFREEAGATG--PFEIHVISMDAYTVDGIKR-LQDKGVTDVIVGFRVPYI
MAV_1061|M.avium_104 RFREEAGKTG--PFEIHVISADAYTPDGIKR-LEDKGVTDVIVGFRIPYI
MAB_2449|M.abscessus_ATCC_1997 KFREEEGTVEK-PFEVHVISVDGFTVDGVKR-LEDKGVTDVIVGFRIPYI
MUL_4083|M.ulcerans_Agy99 QLRAEYGRAEQ-PFEFQAVCIDKFGVQGHRE-LAEAGVTDNIVIPWVFDG
MSMEG_2811|M.smegmatis_MC2_155 RLCARAGRDVD--DLHCAVALQAPAVGDIPA-LVDLGVDELVIVESPP--
MLBr_00269|M.leprae_Br4923 SYDPDPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKVADALPIEQVAKR
:. . : * : :
Mb0965c|M.bovis_AF2122/97 MGPDTEPLQTKIRN-----------------------LEMFAENVIAKV-
Rv0940c|M.tuberculosis_H37Rv MGPDTEPLQTKIRN-----------------------LEMFAENVIAKV-
MMAR_4568|M.marinum_M KGPDTEPLQTKIAH-----------------------LEKFAEKIISKV-
Mflv_1850|M.gilvum_PYR-GCK TGQDTEPLETKIRN-----------------------LEMFAENVIAKA-
Mvan_4885|M.vanbaalenii_PYR-1 QGQDTEPLDAKIRN-----------------------LEMFAENVIAKL-
TH_2326|M.thermoresistible__bu KGPDTEPLADKIRN-----------------------LEMFAENVIAKL-
MAV_1061|M.avium_104 KGKDTEPLDTKIRN-----------------------LEMFAENVIAKV-
MAB_2449|M.abscessus_ATCC_1997 VGPDTEPLDTKIRN-----------------------LESFAEHVISKVN
MUL_4083|M.ulcerans_Agy99 LGFD-APLGKKQDS-----------------------MKRFADTYIHSGW
MSMEG_2811|M.smegmatis_MC2_155 -----QDPDVAADW-----------------------VDALADRWITCI-
MLBr_00269|M.leprae_Br4923 WIVVSDPDEAVARVGQYVTWGLNHLVFHAPGHNQRRFLELFEKDLAPRLR
:. : .
Mb0965c|M.bovis_AF2122/97 ---
Rv0940c|M.tuberculosis_H37Rv ---
MMAR_4568|M.marinum_M ---
Mflv_1850|M.gilvum_PYR-GCK ---
Mvan_4885|M.vanbaalenii_PYR-1 ---
TH_2326|M.thermoresistible__bu ---
MAV_1061|M.avium_104 ---
MAB_2449|M.abscessus_ATCC_1997 P--
MUL_4083|M.ulcerans_Agy99 QAA
MSMEG_2811|M.smegmatis_MC2_155 ---
MLBr_00269|M.leprae_Br4923 RLG