For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKTVVRVALSSIVGLVAFGLIVFIPAGTFDYWQAWVFMAVFAVATLVPSIYLGRTNPAALQRRMHAGPRA ESRGIQKVIITAAFVALFAMVALSALDHRFGWSSVPTAVCLIGDLLIATGLGIAMLVVIQNGYAAATITV ESDQKVVSTGLYSLVRHPMYVGNVILMLGIPLALGSYWGLVIVPPSVLVLAFRILDEESALVDQLDGYRD YMQQVRYRLLPRVW
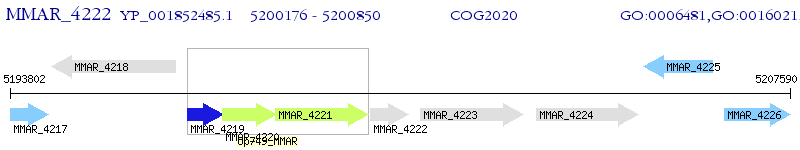
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4222 | - | - | 100% (224) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1248c | - | 1e-81 | 63.18% (220) | integral membrane protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv1216c | - | 1e-81 | 63.18% (220) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4689c | - | 1e-77 | 58.48% (224) | hypothetical protein MAB_4689c |
| M. avium 104 | MAV_1360 | - | 3e-85 | 66.52% (224) | isoprenylcysteine carboxyl methyltransferase (icmt) family |
| M. smegmatis MC2 155 | MSMEG_5077 | - | 6e-67 | 52.23% (224) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0548 | - | 1e-61 | 50.91% (220) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4523 | - | 1e-122 | 99.11% (224) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4501 | - | 2e-70 | 58.48% (224) | isoprenylcysteine carboxyl methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5077|M.smegmatis_MC2_155 MKLL------LQAVASGVFGLAFFAVALFWPAGTFDYWQAWVFIAVFLAG
Mvan_4501|M.vanbaalenii_PYR-1 MKLV------AQTLASSVLGAAYFVAVLFWPAGTFDYWQAWVFLAVFIVT
MMAR_4222|M.marinum_M MKTV------VRVALSSIVGLVAFGLIVFIPAGTFDYWQAWVFMAVFAVA
MUL_4523|M.ulcerans_Agy99 MKTV------VRVALSSIVGLVAFGLIVFIPAGTFDYWQARVFMAVFAVA
MAV_1360|M.avium_104 MKTG------LRVTATSVWGILSWILILLLPAGTLHYWQAWVFIAVFTVA
Mb1248c|M.bovis_AF2122/97 MHIG------LKIFIWGVLGLVVFGALLFGPAGTFDYWQAWVFLAAFVST
Rv1216c|M.tuberculosis_H37Rv MHIG------LKIFIWGVLGLVVFGALLFGPAGTFDYWQAWVFLAAFVST
MAB_4689c|M.abscessus_ATCC_199 MKTGGLVKAIAKVVAFGVVELAVFGLVLFLLAGTVDYWQVWTFLVVFALS
TH_0548|M.thermoresistible__bu VY--------LRTTIYSVFGMVLFGALMFGPAGTLGYWQGWALLAVLVAF
: : .: : :: ***. *** .::..:
MSMEG_5077|M.smegmatis_MC2_155 TLIPSFYLAVTDPVTLQRRLHAGPTAETRSAQKVAAVALVVAVLAVLIVS
Mvan_4501|M.vanbaalenii_PYR-1 TLGPTFFLAVRHPDALARRMKAGPTAETRPAQRLIITLTVVLVTATFVLS
MMAR_4222|M.marinum_M TLVPSIYLGRTNPAALQRRMHAGPRAESRGIQKVIITAAFVALFAMVALS
MUL_4523|M.ulcerans_Agy99 TLVPSIYLGRTNPAALQRRMHAGPRAESRGIQKVIITAAFVALFAMVALS
MAV_1360|M.avium_104 TIVPAAYLARTNPAALQRRMRAGPRAEPRTAQKFIITGSFLSLFAAMVFS
Mb1248c|M.bovis_AF2122/97 TIGPTIYLARNDPAALQRRMRSGPLAEGRTIQKFIVIGAFLGFFAMMVLS
Rv1216c|M.tuberculosis_H37Rv TIGPTIYLARNDPAALQRRMRSGPLAEGRTIQKFIVIGAFLGFFAMMVLS
MAB_4689c|M.abscessus_ATCC_199 TWIPTVYLQRADPAAHQRRKRAGPVAETRSLQKVLIGSWYFSLAAMVVIS
TH_0548|M.thermoresistible__bu TVPYTVYLAVKMPDTLRRRLRSGPTAESRPAQRAAILVVQLAALGMLVLG
* : :* * : ** ::** ** * *: . . . ..
MSMEG_5077|M.smegmatis_MC2_155 ALDRRFGWSSVPAWLALAADVVVVVSLVLSQVVIFQNRFAAATIRVEAEQ
Mvan_4501|M.vanbaalenii_PYR-1 ALDHRFGWSSAPAWVVVVGNTLVALGLSGAQLVVVQNHYAAATVRVEAGQ
MMAR_4222|M.marinum_M ALDHRFGWSSVPTAVCLIGDLLIATGLGIAMLVVIQNGYAAATITVESDQ
MUL_4523|M.ulcerans_Agy99 ALDHRFGWSSVPTAVCLIGDLLIATGLGIAMLVVIQNGYAAATITVECDQ
MAV_1360|M.avium_104 ALDHRFGWSSVPAWLSLLGDVLVATGLGIAMLVVIQNSYAAATVTVEAGQ
Mb1248c|M.bovis_AF2122/97 ACDHRYGWSSVPAAVCVIGDVLVMTGLGIAMLVVIQNRYAASTVRVEAGQ
Rv1216c|M.tuberculosis_H37Rv ACDHRYGWSSVPAAVCVIGDVLVMTGLGIAMLVVIQNRYAASTVRVEAGQ
MAB_4689c|M.abscessus_ATCC_199 ALDHRFGWSSVPPAICAAGDVLVAVGLGVTSLVVVQNGFAASTVRVETGQ
TH_0548|M.thermoresistible__bu GLDHRLGWSRAPVWVCVVGLVLTAAGLAITMGSVLQNAWAAATVTTESDQ
. *:* *** .* : . : .* : :.** :**:*: .* *
MSMEG_5077|M.smegmatis_MC2_155 QVISTGMYGWVRHPMYSWALVMILASPLALGSFWALLAATAGLPVLVFRI
Mvan_4501|M.vanbaalenii_PYR-1 PLVSTGLYGLVRHPMYTGTVVMMLGTPLALGSLWGLLGVAASAPVLVARI
MMAR_4222|M.marinum_M KVVSTGLYSLVRHPMYVGNVILMLGIPLALGSYWGLVIVPPSVLVLAFRI
MUL_4523|M.ulcerans_Agy99 KVVSTGLYSLVRHPMYVGNVILMLGIPLALGSYWGLVIVPPSVLVLAFRI
MAV_1360|M.avium_104 TVVSSGLYRFVRHPMYTGNVIMMIGIPLALGSYWGLLFVVPGTALLALRI
Mb1248c|M.bovis_AF2122/97 ILASDGLYKIVRHPMYAGNVVMMTGIPLALGSYWAMFILVPGTLVLVFRI
Rv1216c|M.tuberculosis_H37Rv ILASDGLYKIVRHPMYAGNVVMMTGIPLALGSYWAMFILVPGTLVLVFRI
MAB_4689c|M.abscessus_ATCC_199 TLVTTGLYGLVRHPMYTGNFITIVGFPLALGSYWGLACVVPSLLVLIVRI
TH_0548|M.thermoresistible__bu QVVSTGLYGVVRHPLYAGSVVLFLGMPLALGSYWALALVVVAVAGLVVRI
: : *:* ****:* .: : . ****** *.: . * **
MSMEG_5077|M.smegmatis_MC2_155 LDEEKMLVAELRGYDEYRQKIRYRLVPGVW
Mvan_4501|M.vanbaalenii_PYR-1 LDEEKMLTAELAGYREYRRQVRYRLVPGVW
MMAR_4222|M.marinum_M LDEESALVDQLDGYRDYMQQVRYRLLPRVW
MUL_4523|M.ulcerans_Agy99 LDEESALVDQLDGYRDYMQQVRYRLLPRVW
MAV_1360|M.avium_104 FDEEKLLLQELPGYPDYTQKVRYRLAPHLW
Mb1248c|M.bovis_AF2122/97 LDEEKLLTQELSGYREYRQLVRYRLVPYVW
Rv1216c|M.tuberculosis_H37Rv LDEEKLLTQELSGYREYRQLVRYRLVPYVW
MAB_4689c|M.abscessus_ATCC_199 VDEEKLLVEELDGYRAYTRAARYRLMPYVW
TH_0548|M.thermoresistible__bu LDEEALLRTELAGYPDYTRAVPYRLVPYVW
.*** * :* ** * : *** * :*