For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEAVGLSDELGPVDYLMHRGEANPRTRSGIMALELLDTTPDWERFRTRFENASRRVLRLRQKVVVPTLPT AAPRWVVDPDFNLDFHVRRVRVSEPGTLRDVFDLAEVILQSPLDISRPLWTATLVEGLADGKAATLLHVS HAVTDGVGGVEMFAQIYDLERDPAPRPTPPLPIPQDLSPNDLMREGIGHLPIALVGGVAGAIAGAVSVAG RAVLDPAATVSGIVGYALSSVRVMNRAAEPSPLLRRRSLTTRTEAIDIRLSELHKAAKAGGGSINDAYLA GLCGALRRYHEALGMPISTLPMAVPVNLRADADGAGGNRFTGVNLAAPVGTADPVARMKKIRAQMTQRRD EPAMNVVGSMAPVLSVLPTAVLEGISSSVIGSDVQASNVPVYPGDTFVAGAKILRQYGIGPLPGVAMMVV LISRGGWATITVRYDRASVRHDELFARCLVEGFDEILALAGDPAPRSIPASFTTVSGSESRSVSNS*
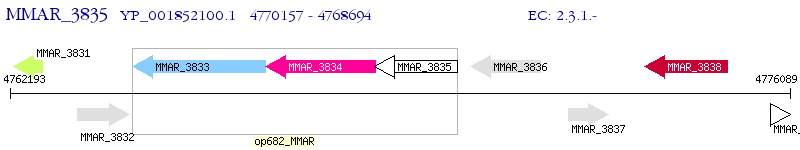
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3835 | - | - | 100% (487) | hypothetical protein MMAR_3835 |
| M. marinum M | MMAR_2649 | - | 6e-74 | 37.05% (448) | hypothetical protein MMAR_2649 |
| M. marinum M | MMAR_1311 | - | 9e-30 | 27.37% (464) | hypothetical protein MMAR_1311 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2509c | - | 0.0 | 87.24% (486) | hypothetical protein Mb2509c |
| M. gilvum PYR-GCK | Mflv_2563 | - | 0.0 | 68.20% (478) | hypothetical protein Mflv_2563 |
| M. tuberculosis H37Rv | Rv2484c | - | 0.0 | 87.04% (486) | hypothetical protein Rv2484c |
| M. leprae Br4923 | MLBr_01244 | - | 0.0 | 74.07% (486) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_4544c | - | 6e-31 | 28.57% (420) | hypothetical protein MAB_4544c |
| M. avium 104 | MAV_1686 | - | 0.0 | 85.15% (485) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. smegmatis MC2 155 | MSMEG_4705 | - | 0.0 | 70.23% (477) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. thermoresistible (build 8) | TH_1039 | - | 0.0 | 70.49% (471) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. ulcerans Agy99 | MUL_3765 | - | 0.0 | 97.97% (493) | hypothetical protein MUL_3765 |
| M. vanbaalenii PYR-1 | Mvan_4076 | - | 0.0 | 68.64% (472) | hypothetical protein Mvan_4076 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2563|M.gilvum_PYR-GCK --MSN--SSGLDAAGLPEELSPLDQILHRGEANPRTRSGIMTIEL-LDTT
Mvan_4076|M.vanbaalenii_PYR-1 --MSN--EPELDAAGLPEELSPLDQILHRGEGNPRTRSGIMTLEL-LDTT
TH_1039|M.thermoresistible__bu --MSD--VPELDAAGLPEELSGLDQILHRGEANPRTRSGIMAVEL-LDTT
MSMEG_4705|M.smegmatis_MC2_155 --MSY--VPERDAAGLPEELSAVDQLLHRGEANPRTRSGIMGVEL-LDTT
MMAR_3835|M.marinum_M ---------MAEAVGLSDELGPVDYLMHRGEANPRTRSGIMALEL-LDTT
MUL_3765|M.ulcerans_Agy99 --MAEWVVIMAEAVGLSDELGPVDYLMHRGEANPRTCSGIMALEL-LDTT
Mb2509c|M.bovis_AF2122/97 --MAE----SGESPRLSDELGPVDYLMHRGEANPRTRSGIMALEL-LDGT
Rv2484c|M.tuberculosis_H37Rv --MAE----SGESPRLSDELGPVDYLMHRGEANPRTRSGIMALEL-LDGT
MAV_1686|M.avium_104 MTMTE----SVETIKLSDELGPVDYLMHRGEANPRTRSGIMALEL-LDTT
MLBr_01244|M.leprae_Br4923 --MAD----SVEGIGPFDELGALDYLLHRGEANPRTRAGIMAVEL-LDTT
MAB_4544c|M.abscessus_ATCC_199 --------------MPFMPVSDSMFLIGETREHPFHVGGLQLFKPPVDAG
:. :: . . :* .*: .: :*
Mflv_2563|M.gilvum_PYR-GCK PDWDT-FRTRFEHASRKVLRLRQKVVTPTLPTTAPRWVVDPDFNLDFHLR
Mvan_4076|M.vanbaalenii_PYR-1 PDWDT-FRTRFDNASRKVLRLRQKVVTPTLPTAAPRWVVDPDFNLDFHVR
TH_1039|M.thermoresistible__bu PDWDR-FRTRFDHASRRVLRLRQKVVMPTLPTAPARWVVDPDFNLDYHVR
MSMEG_4705|M.smegmatis_MC2_155 PDWER-FRTRFDHASRKVVRLRQKVVMPTLPTAAPRWVVDPDFNLDYHVR
MMAR_3835|M.marinum_M PDWER-FRTRFENASRRVLRLRQKVVVPTLPTAAPRWVVDPDFNLDFHVR
MUL_3765|M.ulcerans_Agy99 PDWER-FRTRFENASRRVLRLRQKVVVPTLPTAAPRWVVDPDFNLDFHVR
Mb2509c|M.bovis_AF2122/97 PDWDR-FRTRFENASRRVLRLRQKVVVPTLPTAAPRWVVDPDFNLDFHVR
Rv2484c|M.tuberculosis_H37Rv PDWDR-FRTRFENASRRVLRLRQKVVVPTLPTAAPRWVVDPDFNLDFHVR
MAV_1686|M.avium_104 PDWQQ-FRARFENASRRVLRLRQKVVVPTLPTAAPRWVIDPDFNLDFHVR
MLBr_01244|M.leprae_Br4923 PDWNR-FRSRIEDVSQRVLRLRQKVVVPTLPTAAPRWVVDPDFELDFHVR
MAB_4544c|M.abscessus_ATCC_199 PDYAEVFYEQLMSTTEVSSDFRKRPGQPLAVMGNLTWIVDDEVDWEYHVR
**: * :: .:. :*:: * *::* :.: ::*:*
Mflv_2563|M.gilvum_PYR-GCK RVRVPEPGTLRQVMDFAEIAAQSPLDISRPLWTATLIEGVQDGRAALMVH
Mvan_4076|M.vanbaalenii_PYR-1 RVRVPQPGTFRQVMDLAEIAAQSPLDISRPLWTATLIEGVENGRAALMVH
TH_1039|M.thermoresistible__bu RVRVPEPGTLRQVFDLAEVDMQSPLDISRPLWTATLVEGLRDGRAATMLH
MSMEG_4705|M.smegmatis_MC2_155 RVRVPGPGTLRQVLDLAEVAMQSPMDITRPLWSATLVEGLADGRAATILH
MMAR_3835|M.marinum_M RVRVSEPGTLRDVFDLAEVILQSPLDISRPLWTATLVEGLADGKAATLLH
MUL_3765|M.ulcerans_Agy99 RVRVSEPGTLRDVFDLAEVILQSPLDISRPLWTATLVEGLADGKAATLLH
Mb2509c|M.bovis_AF2122/97 RVRVSGPATLREVLDLAEVILQSPLDISRPLWTATLVEGMADGRAAMLLH
Rv2484c|M.tuberculosis_H37Rv RVRVSGPATLREVLDLAEVILQSPLDISRPLWTATLVEGMADGRAAMLLH
MAV_1686|M.avium_104 RVRVAEPGTLREVFDLAELILQSPMDISRPLWTATLVDGLADGKAATLLH
MLBr_01244|M.leprae_Br4923 RVRVPDPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLAAGRAAMLLQ
MAB_4544c|M.abscessus_ATCC_199 RSALPRPARVRELLTVTSRWHSSPLDRHRPLWEMHVVEGLSDGRLAVYTK
* :. *. .*::: .:. .**:* **** :::*: *: * :
Mflv_2563|M.gilvum_PYR-GCK LSHAVTDGVGGVEMFANLYDFEREPPPQPVPPMPVPTDLSPNDLMRSGLN
Mvan_4076|M.vanbaalenii_PYR-1 LSHAVTDGVGGVEMFANLYDFERDPPPQSTPPLPIPSDLSPNDLMRAGLN
TH_1039|M.thermoresistible__bu LSHAVTDGVGGVEMFASIYDFERDPPPGKEPPLPIPQDLTPNDLMWEGVG
MSMEG_4705|M.smegmatis_MC2_155 LSHAVSDAVGGVEMFASIYDLERDPEPDTPPPLPIPQDLSPGELMREGIN
MMAR_3835|M.marinum_M VSHAVTDGVGGVEMFAQIYDLERDPAPRPTPPLPIPQDLSPNDLMREGIG
MUL_3765|M.ulcerans_Agy99 VSHAVTDGVGGVEMFAQIYNLERDPAPRPTPPLPIPQDLSPNDLMREGIG
Mb2509c|M.bovis_AF2122/97 VSHAVTDGVGGVEMFAQIYDLERDPPPRSTPPQPIPEDLSPNDLMRRGIN
Rv2484c|M.tuberculosis_H37Rv VSHAVTDGVGGVEMFAQIYDLERDPPPRSTPPQPIPEDLSPNDLMRRGIN
MAV_1686|M.avium_104 VSHAVTDGVGGVQMFAEIYDLERNPPPKPPPPLPVPQDLSPNDLMRQGIN
MLBr_01244|M.leprae_Br4923 ISHAITDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRNDLMLQGIN
MAB_4544c|M.abscessus_ATCC_199 MHHAVIDGVGALRMMMRS--LSDDPDARDCQAVWAPAKRRAKSSIVSTTN
: **: *.**.:.*: :. :* . . * . . : .
Mflv_2563|M.gilvum_PYR-GCK RLPGTIVGGVVGAVAGAAQAVGHVVRDPVERLGSVFDYARSGVRVVGPVA
Mvan_4076|M.vanbaalenii_PYR-1 RLPGTIVGGVRDALFGAAQAVGHVVRDPVSRLGSAFDYAMSGLRVVGPVA
TH_1039|M.thermoresistible__bu RLPRSVVRGVRGLLGDTVRAVGKTLRDPRATVEGVLDYAASSARLVTPVA
MSMEG_4705|M.smegmatis_MC2_155 RLPGSIAGGVLGALGGAARAVTKVVRDPISAVSGVVDYAMSGTRVMAPAA
MMAR_3835|M.marinum_M HLPIALVGGVAGAIAGAVSVAGRAVLDPAATVSGIVGYALSSVRVMNRAA
MUL_3765|M.ulcerans_Agy99 HLPIALVGGVAGAIAGAVSVAGRAVLDSAATVSGIVGYALSSVRVMNRAA
Mb2509c|M.bovis_AF2122/97 HLPIAVVGGVLDALSGAVSMAGRAVLEPVSTVSGILGYARSGIRVLNRAA
Rv2484c|M.tuberculosis_H37Rv HLPIAVVGGVLDALSGAVSMAGRAVLEPVSTVSGILGYARSGIRVLNRAA
MAV_1686|M.avium_104 HLPFAVVGGVVSALSGAVSVAGRAMLEPASTVSGIVGYAMSGMRVLNRAA
MLBr_01244|M.leprae_Br4923 HLPVALAGGVVGGLSGVASVAGRAILRPASTVSGVVGYVRSGIRVLSQAA
MAB_4544c|M.abscessus_ATCC_199 TSAIDLVKGAAGAVRQVVSIPPGLLKYG--------RHALSDPQLVKPLS
. :. *. . : .. : :. *. ::: :
Mflv_2563|M.gilvum_PYR-GCK DPSPILRRRS-LSSRSEAIDIEFSDLHRASKAAEGSINDAYLAGLCGALR
Mvan_4076|M.vanbaalenii_PYR-1 DPSPILRRRS-LSSRSDAIDIEFGDLHRAAKAAGGSINDAYLAGLCGALR
TH_1039|M.thermoresistible__bu EPSPILRRRS-LASRTGAIDIRFEDLRKAAKAGGGSINDAYLTGLCGALR
MSMEG_4705|M.smegmatis_MC2_155 EPSPLLRRRS-LASRSEAIDIPFGDFHRAAKAAGGSINDAYLAGVCGALR
MMAR_3835|M.marinum_M EPSPLLRRRS-LTTRTEAIDIRLSELHKAAKAGGGSINDAYLAGLCGALR
MUL_3765|M.ulcerans_Agy99 EPSPLLRRRS-LTTRTEAIDIRLSELHKAAKAGGGSINDAYLAGLCGALR
Mb2509c|M.bovis_AF2122/97 EPSPLLRRRS-LTTRTEAIDIRLADLHKAAKAGGGSINDAYLAGLCGALR
Rv2484c|M.tuberculosis_H37Rv EPSPLLRRRS-LTTRTEAIDIRLADLHKAAKAGGGSINDAYLAGLCGALR
MAV_1686|M.avium_104 EPSPLLRRRS-LATRSEAIDIPLADLHKAAKAGGGSINDAYLAGLCGALR
MLBr_01244|M.leprae_Br4923 EPSPLLRQRS-LATRTEAIEIQLSDLHKAAKAGDGSINDAYLAGLVGALR
MAB_4544c|M.abscessus_ATCC_199 APHTMLNVSIGGARRFAAQTWSMARIKEAGKALGGTVNDVVLAMCGGALR
* .:*. : * * : ::.*.** *::**. *: ****
Mflv_2563|M.gilvum_PYR-GCK LYHESMG-VPVDTLPMAVPVNLRSDADPAGGNRFVGVN------LAAPVG
Mvan_4076|M.vanbaalenii_PYR-1 LYHEAKG-VPIDTLPMAVPVNLRSEADPAGGNRFVGVN------LAAPIG
TH_1039|M.thermoresistible__bu LYHDALG-VPVDTLPMAVPVNLRSDADPAGGNRFTGVN------LAAPIG
MSMEG_4705|M.smegmatis_MC2_155 LYHDAKG-VPIDSLPMAVPVNLRAEDDPAGGNRFAGVN------LSAPIG
MMAR_3835|M.marinum_M RYHEALG-MPISTLPMAVPVNLRADADGAGGNRFTGVN------LAAPVG
MUL_3765|M.ulcerans_Agy99 RYHEALG-MPISTLPMAVPVNLRADADGAGGNRFTGVNRFTGVNLAAPVG
Mb2509c|M.bovis_AF2122/97 RYHEALG-VPISTLPMAVPVNLRAEGDAAGGNQFTGVN------LAAPVG
Rv2484c|M.tuberculosis_H37Rv RYHEALG-VPISTLPMAVPVNLRAEGDAAGGNQFTGVN------LAAPVG
MAV_1686|M.avium_104 RYHEAFG-VPISTLPMAVPVNLRAEADTAGGNRFTGVN------LAAPIG
MLBr_01244|M.leprae_Br4923 RYHEALG-VSISTLPMAVPVNVRTEADVVGSNRFVGVT------LAAPLG
MAB_4544c|M.abscessus_ATCC_199 TYLEEQNALPDKPLVAMCPVSIRAEGDQNAGNSITALL------ANLATD
* : . :. ..* **.:*:: * ..* :..: . .
Mflv_2563|M.gilvum_PYR-GCK LSDPAMRIKEIRAQMRSKR-----DERALDVVGAIAPLVSLLPDSVLETM
Mvan_4076|M.vanbaalenii_PYR-1 VSDPEVRIQDIRSQMTRKR-----DERAMDMVGAIAPVVSLLPDTVLESM
TH_1039|M.thermoresistible__bu IVDPVDRMQDVRAQMTRKR-----EERALDLLGMVAPVGSLLPDTVLEAL
MSMEG_4705|M.smegmatis_MC2_155 IEDAEQRIKSVRSQMTSLR-----EERAIDVVGAIAPVLSLLPDTILESV
MMAR_3835|M.marinum_M TADPVARMKKIRAQMTQRR-----DEPAMNVVGSMAPVLSVLPTAVLEGI
MUL_3765|M.ulcerans_Agy99 TADPVARMKKIRAQMTQRR-----DEPAMNVVGSMAPVLSVLPTAVLEGI
Mb2509c|M.bovis_AF2122/97 TIDPVARMKKIRAQMTQRR-----DEPAMNIIGSIAPVLSVLPTAVLEGI
Rv2484c|M.tuberculosis_H37Rv TIDPVARMKKIRAQMTQRR-----DEPAMNIIGSIAPVLSVLPTAVLEGI
MAV_1686|M.avium_104 TVDPVSRMKKIRAQMTQRR-----DEPAMNIIGSLAPVLSVLPTAVLEGI
MLBr_01244|M.leprae_Br4923 TNDPAARMQKIRSQMTQRR-----DEPAMNIIGSLAPLMTVLPASVLDFI
MAB_4544c|M.abscessus_ATCC_199 KPDPLERFSAIRDSVQAAKSVLSEMTPLQRMLVGALNGAPVLTGAVPGLV
*. *:. :* .: : :: .:*. :: :
Mflv_2563|M.gilvum_PYR-GCK AGSIVNSDVQASNVPVYAGDTFIAGAKVLRQYGLG-PLPGVAMMVVLISR
Mvan_4076|M.vanbaalenii_PYR-1 AGSFVNSDVQASNVPVYAGDTFIAGAKVLRQYGLG-PLPGVAMMVVLISR
TH_1039|M.thermoresistible__bu AGTVVNSDVQASNVPVILEDTYIAGAKVLRQYGLG-PLPGVAMMVVLVSR
MSMEG_4705|M.smegmatis_MC2_155 AGSVVNSDVQASNVPIYPGDTFLAGAKILRHYGLG-PLPGVAMMVVLISR
MMAR_3835|M.marinum_M SSSVIGSDVQASNVPVYPGDTFVAGAKILRQYGIG-PLPGVAMMVVLISR
MUL_3765|M.ulcerans_Agy99 SSSVIGSDVQASNVPVYPGDTFVAGAKILRQYGIG-PLPGVAMMVVLISR
Mb2509c|M.bovis_AF2122/97 TGSVIGSDVQASNVPVYPGDTYLAGAKILRQYGIG-PLPGVAMMVVLISR
Rv2484c|M.tuberculosis_H37Rv TGSVIGSDVQASNVPVYPGDTYLAGAKILRQYGIG-PLPGVAMMVVLISR
MAV_1686|M.avium_104 TGSVIGSDVQASNVPVFPGDTYIAGAKVLRQYGIG-PLPGVAMMVVLISR
MLBr_01244|M.leprae_Br4923 VDSVASSDVNASNIPAYPGDTYFAGAKILRQYGIG-PRPGVAMMAVLMSR
MAB_4544c|M.abscessus_ATCC_199 DRTAPAFNVIISNVPGPRTDMFWNGFEMDGCYPASIPIDGVALNITCTSS
: :* **:* * : * :: * . * ***: . *
Mflv_2563|M.gilvum_PYR-GCK SGYCTISTRYDRASITDPGLLAQCLLDGFDEVLALGGD--GRAVPATFSR
Mvan_4076|M.vanbaalenii_PYR-1 SGYCTVTARYDRAAITDPDLLAQCLLAGFDEVLALGGD--GRAVPATFTA
TH_1039|M.thermoresistible__bu SGYFTVTARYDRAAITDARLFAHCLLAGFNEVLALGGD--GRAEPASFEV
MSMEG_4705|M.smegmatis_MC2_155 GGYITVTVRYDRAAITDGALFARCLQQGFDEVLALGGE--VRCTPASFDP
MMAR_3835|M.marinum_M GGWATITVRYDRASVRHDELFARCLVEGFDEILALAGDPAPRSIPASFTT
MUL_3765|M.ulcerans_Agy99 GGWATITVRYDRASVRHDELFARCLVEGFDEILVLAGDPAPRSIPASFTT
Mb2509c|M.bovis_AF2122/97 GGWCTVTVRYDRASVRNDELFAQCLQAGFDEILALAGDPAPRVLPASFDT
Rv2484c|M.tuberculosis_H37Rv GGWCTVTVRYDRASVRNDELFAQCLQAGFDEILALAGGPAPRVLPASFDT
MAV_1686|M.avium_104 GGWCTITVRYDRASVRDEALFAQCLLEGFDEILALAGDGAPRAVPASFTE
MLBr_01244|M.leprae_Br4923 GGFCTVTVRYDRASVKSEALFARCLLEGFDEVLALAGDPTPHAVPASFAA
MAB_4544c|M.abscessus_ATCC_199 GSSMHFGLTGCRTSVPHLQRILAHLEDALTQLEESVA-------------
.. . *::: : * .: :: .
Mflv_2563|M.gilvum_PYR-GCK VGAQEK---------------
Mvan_4076|M.vanbaalenii_PYR-1 QTTPPVDSARLNGSGPQ----
TH_1039|M.thermoresistible__bu DPARLEALMSNAVVPSESAMR
MSMEG_4705|M.smegmatis_MC2_155 EPGTNIGGAQ-----------
MMAR_3835|M.marinum_M V-SGSESRSVSNS--------
MUL_3765|M.ulcerans_Agy99 V-SGSESRSVSNS--------
Mb2509c|M.bovis_AF2122/97 QGAGSVPRSVSGS--------
Rv2484c|M.tuberculosis_H37Rv QGAGSVPRSVSGS--------
MAV_1686|M.avium_104 S--AVSARSASGS--------
MLBr_01244|M.leprae_Br4923 RSSGSPAGWLSSS--------
MAB_4544c|M.abscessus_ATCC_199 ---------------------