For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGRYVENLGVRIWTQSFGSRADKAILLISGAMAPAIFWNSNFCERLADLGHFVLRFDSRDIGDSTHFPPA RDADADPPYTIDEMVDDVRSILADYDLNTVVLIGHSLGSTVAQLFAVKYPERVEKLFLMSSPIIATGNNT YAETDSETLAAMWQVLMSNKMYPDYKRGKDEFFKVWRYLNGSFELDTDLADEYTKRLYETEVIEPAYNHT KIQVGIDDTWTELSELEIPIHFIYGEKDQLASNVGNIQILANSLPNAHLTVLKGAGHMYFHRRIWQQILS VLTEALA
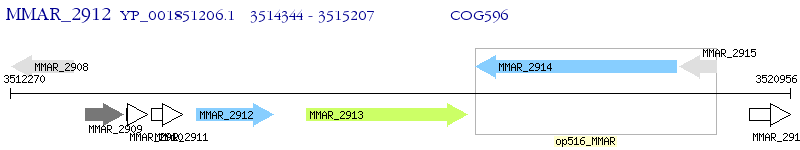
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2912 | - | - | 100% (287) | hydrolase or acyltransferase |
| M. marinum M | MMAR_0981 | lipG1 | 1e-17 | 24.49% (294) | lipase/esterase LipG1 |
| M. marinum M | MMAR_0983 | lipG2 | 1e-13 | 24.41% (295) | lipase/esterase LipG2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0665c | lipG | 2e-14 | 21.96% (296) | lipase/esterase LipG |
| M. gilvum PYR-GCK | Mflv_5112 | - | 4e-16 | 25.67% (300) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0646c | lipG | 2e-14 | 21.96% (296) | lipase/esterase LipG |
| M. leprae Br4923 | MLBr_01899 | lipG | 6e-15 | 29.66% (145) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3880 | - | 6e-18 | 24.12% (311) | lipase/esterase LipG |
| M. avium 104 | MAV_4515 | - | 2e-15 | 22.67% (300) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_1352 | - | 2e-18 | 24.24% (297) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_1861 | lipG | 2e-16 | 25.84% (298) | PROBABLE LIPASE/ESTERASE LIPG |
| M. ulcerans Agy99 | MUL_0733 | lipG1 | 5e-17 | 24.15% (294) | lipase/esterase LipG1 |
| M. vanbaalenii PYR-1 | Mvan_1240 | - | 2e-16 | 25.17% (298) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1352|M.smegmatis_MC2_155 -----MQTRTGLARAGDAD---------HPIDLFYEDMGDPGDPAVLLIM
TH_1861|M.thermoresistible__bu -----MQIRTGFANVDGSP---------EPVDLYYEDLGDPADPPVLLIM
Mflv_5112|M.gilvum_PYR-GCK -----MQVRTGTARSGE-------------LEIHYEDMGDPNDPAVLLIM
Mvan_1240|M.vanbaalenii_PYR-1 -----MQIRSGTAKSGD-------------LDIFYEDMGDPNDPAVLLIM
Mb0665c|M.bovis_AF2122/97 -----MDIRSGTAVS-------------GDVKLYYEDMGDLDHPPVLLIM
Rv0646c|M.tuberculosis_H37Rv -----MDIRSGTAVS-------------GDVKLYYEDMGDLDHPPVLLIM
MAV_4515|M.avium_104 ----MVQVRTGHAIS-------------GDLKLYYEDMGDIDDPPVLLIM
MLBr_01899|M.leprae_Br4923 -----MEVRSGYATS-------------GDLKLYYEDMGNVDDPPVLLIM
MUL_0733|M.ulcerans_Agy99 -----MEIRTGYAQSGVGSGATSERQPAGDLKLYYEDMGDINAPPVLLIM
MAB_3880|M.abscessus_ATCC_1997 MSTTAREISEMTASTAT----------VGDIELCYEEFGNPADPAVLLIM
MMAR_2912|M.marinum_M -------MGRYVENLG--------------VRIWTQSFGSRADKAILLIS
: : :.:*. .:***
MSMEG_1352|M.smegmatis_MC2_155 GLGAQMLMWRTEFCEKLVDQGLRVIRFDNRDVGLSTKLS---GQRVDSPL
TH_1861|M.thermoresistible__bu GLGAQLILWRDGFCRKLVERGLRVIRFDNRDVGLSSKLD---GRRERTPL
Mflv_5112|M.gilvum_PYR-GCK GLGAQLLLWRKGFCEKLINQGLRVIRFDNRDVGLSSKLT---GHHAGAPL
Mvan_1240|M.vanbaalenii_PYR-1 GLGAQLLLWRNGFCEKLINQGLRVIRFDNRDVGLSSKLH---HHRSGAPL
Mb0665c|M.bovis_AF2122/97 GLGAQMLLWRTDFCARLVAKGLRVIRYDNRDVGLSTKTE---RHRPGQPL
Rv0646c|M.tuberculosis_H37Rv GLGAQMLLWRTDFCARLVAKGLRVIRYDNRDVGLSTKTE---RHRPGQPL
MAV_4515|M.avium_104 GLGAQLLLWRTAFCEKLVGRGLRVIRYDNRDVGLSSKTE---HRSSGQPL
MLBr_01899|M.leprae_Br4923 GLGAQLVLWRTAFCEKLVAQGLRVVRYDNRDVGLSSRTDSTEQPCPSQPL
MUL_0733|M.ulcerans_Agy99 GLGAQLLLWRTGFCQKPVDQGLRVIRYDNRDVGLSTKME---QPSARQPL
MAB_3880|M.abscessus_ATCC_1997 GIGAQMVFWRPEFCQQLADQGYRVIRFDNRDCGLSTKLDG--VRAGGGSL
MMAR_2912|M.marinum_M GAMAPAIFWNSNFCERLADLGHFVLRFDSRDIGDSTHFP-----------
* * ::*. ** : * *:*:*.** * *::
MSMEG_1352|M.smegmatis_MC2_155 ALRMARSFLGKRSP-AVYTLEDMADDAAALLDHLEIDRAHIVGGSMGGMI
TH_1861|M.thermoresistible__bu PWRMLRSFAGLPSP-AVYTLEDMADDAAGLLDHLQIDAAHIVGASMGGMV
Mflv_5112|M.gilvum_PYR-GCK LPRMARSLAGLPSP-AAYTLEDMADDAAALLDHLDIDRAHVVGGSMGGMI
Mvan_1240|M.vanbaalenii_PYR-1 LPRMARSFVGRPSP-AAYTLEDMADDAAALLDHLGIDTAHVVGGSMGGMI
Mb0665c|M.bovis_AF2122/97 ATRLVRSWLGLPSQ-AAYTLEDMAADAAALLDHLDVKHAHVVGASMGGMI
Rv0646c|M.tuberculosis_H37Rv ATRLVRSWLGLPSQ-AAYTLEDMAADAAALLDHLDVKHAHVVGASMGGMI
MAV_4515|M.avium_104 VTRLLRSWLGLPSQ-SAYRLEDMADDAAAVLDHLGIDDAHIVGASMGGMI
MLBr_01899|M.leprae_Br4923 TARLIRFWLGQRNA-CAYTLEDMTDDAVALLDHLSIERAHIVGASMGGMI
MUL_0733|M.ulcerans_Agy99 MPQLIRSWLGRRSQ-TPYTLEDMADDAAALLDHLDIERAHIVGASMGGMI
MAB_3880|M.abscessus_ATCC_1997 IPSMAKFLAGVKITGTAYTLVDMAGDAAGLLDHLGIEQAHIVGASMGGMI
MMAR_2912|M.marinum_M -----PARDADADP--PYTIDEMVDDVRSILADYDLNTVVLIGHSLGSTV
. * : :*. *. .:* . :. . ::* *:*. :
MSMEG_1352|M.smegmatis_MC2_155 AQVFASQHAARTETLGVIFSSNNQALLPPPGIRQLLSLITGPPADASRDA
TH_1861|M.thermoresistible__bu AQVFAGRHAHRTRSLGVIFSSNNRPFLPPPGPRQLLSIITGPAPDAPRDE
Mflv_5112|M.gilvum_PYR-GCK AQVFAARHHPRTKTLGVIFSSNNQAALPPPGPRQLAAILQRPK-GSSRDE
Mvan_1240|M.vanbaalenii_PYR-1 AQVFAARHRARTGALGVIFSSNNQAALPPPGPRQLAAILQRPK-GSDREA
Mb0665c|M.bovis_AF2122/97 AQIFAARFAQRTKTLAVIFSSNNHRFLPPPAPRALLALLTGPPPDSPRDV
Rv0646c|M.tuberculosis_H37Rv AQIFAARFAQRTKTLAVIFSSNNHRFLPPPAPRALLALLTGPPPDSPRDV
MAV_4515|M.avium_104 AQIFAARFRQRTKTLAVIFSSNNSALLPPPAPRALLALLKGPPPDSPREV
MLBr_01899|M.leprae_Br4923 AQIFAARFPTRTRSLAVFFSSNNRPFLPPPAPRALLALLTGPPTGSRRDV
MUL_0733|M.ulcerans_Agy99 AQIFAARFAERTNSLAVIFSSNNRPFLPPPAPRALLAVLNGPPPGSPREV
MAB_3880|M.abscessus_ATCC_1997 AQVFAAEHPDRTATVSIIMSSNNQPFLPPPGPRQLMALLKPPPEGASREQ
MMAR_2912|M.marinum_M AQLFAVKYPERVEKLFLMSSPIIATGNNTYAETDSETLAAMWQVLMSNKM
**:** .. *. : :: *. . . :: ..
MSMEG_1352|M.smegmatis_MC2_155 VLDNAVRLSRLNGSPAYPRSEEEIRAEAAELYDRAYYPAGIAR----HFA
TH_1861|M.thermoresistible__bu VIENAVKVGRLNGSPAYPVPDEQLRADAATAFDRSYHPVGVAR----QFD
Mflv_5112|M.gilvum_PYR-GCK IVDNAVRVSRIIGSPGYPAPLDRLRTDAIEGFDRSYYPAGVGR----HFA
Mvan_1240|M.vanbaalenii_PYR-1 IIENAVRVSRIIGSPGYPAAEERLRADAIEGYDRSYYPAGVGR----HFA
Mb0665c|M.bovis_AF2122/97 IVDNAVRVSKIIGSPAYPIPEDQVRAEAAESYDRNFHPWGIAQ----QFS
Rv0646c|M.tuberculosis_H37Rv IVDNAVRVSKIIGSPAYPIPEDQVRAEAAESYDRNFHPWGIAQ----QFS
MAV_4515|M.avium_104 IIDNAVRVGRIIGSTRYRVPEEQARAEAAEGYDRNYYPQGVAR----HFS
MLBr_01899|M.leprae_Br4923 VVDNVVRVTKITGSPLYRMPEEQVRTNAAEIYDRSFYPLGVSR----QFS
MUL_0733|M.ulcerans_Agy99 IIDNVVRVTRITGSPGYQPTEEWIRADAAENYDRSYYPWGIPR----QFS
MAB_3880|M.abscessus_ATCC_1997 IIANSVQVGRIIGSPKYRQSQSKSYLHAAEYYDRSYYPKGFSR----QFA
MMAR_2912|M.marinum_M YPDYKRGKDEFFKVWRYLNGSFELDTDLADEYTKRLYETEVIEPAYNHTK
.: * . : : : . . :
MSMEG_1352|M.smegmatis_MC2_155 AILGSGSLRHHNERVTAPTVVIHGRADRLMPPSG-GRAIANAIRGARMVL
TH_1861|M.thermoresistible__bu AILGSGSLRRYNRRTTSPTVVIHGRADKLMRPTG-GRAVARAIPGARLTL
Mflv_5112|M.gilvum_PYR-GCK AILGSGSLRRYDRQITAPTVVIHGRADKLMRPSG-GRAIARAIPGARLVL
Mvan_1240|M.vanbaalenii_PYR-1 AILGSGSLRRYDRQITAPTVVIHGTADKLMRPSG-GRAIARAIPRARLVL
Mb0665c|M.bovis_AF2122/97 AILGSGSLLRYDRRIVAPTVVIHGRADKLMRPFG-GRAVARAINGARLVL
Rv0646c|M.tuberculosis_H37Rv AILGSGSLLRYDRRIVAPTVVIHGRADKLMRPFG-GRAVARAINGARLVL
MAV_4515|M.avium_104 AVLGSGSLRRYNRRTTAPTVVIHGRADKLMRPFG-GRAVAGAIDGARLVL
MLBr_01899|M.leprae_Br4923 AILGSGSLLHYNQRIIAPTVVIHGRADKLVRPSS-GRAVARAITGARLVL
MUL_0733|M.ulcerans_Agy99 AVLGSGSLLGYNRRTVAPTVVIHGRADKLMRPFG-GRAVAKAINGARMVE
MAB_3880|M.abscessus_ATCC_1997 AIMGTGSLAPFDNRITAPALVLHGKADKLMRPSG-ARAIARAIPGSRLAL
MMAR_2912|M.marinum_M IQVGIDDTWTELSELEIPIHFIYGEKDQLASNVGNIQILANSLPNAHLTV
:* .. . * .::* *:* . : :* :: :::.
MSMEG_1352|M.smegmatis_MC2_155 IDGMGHE-LPEPVWDEVIGELKTNFAVVG-------------
TH_1861|M.thermoresistible__bu FDGMGHD-LPEPLWDEIVAELDKNITAGR-------------
Mflv_5112|M.gilvum_PYR-GCK FDGMGHD-LPEPLWDDIVGELKTTFAEAP-------------
Mvan_1240|M.vanbaalenii_PYR-1 FDGMGHE-LPEPLWDDIVGELKTTFAEIS-------------
Mb0665c|M.bovis_AF2122/97 IDGMGHD-LPRQLWDRVIGELTRNFSEAG-------------
Rv0646c|M.tuberculosis_H37Rv IDGMGHD-LPRQLWDRVIGELTRNFSEAG-------------
MAV_4515|M.avium_104 FDGMGHD-LPQQLWDQVIGVLANNFAKAS-------------
MLBr_01899|M.leprae_Br4923 FDGMGHD-LPQQLWDQAVGVLMSNFAKAS-------------
MUL_0733|M.ulcerans_Agy99 FDGMGHD-LPRQLWDQVIGVLTSNFEQAE-------------
MAB_3880|M.abscessus_ATCC_1997 IDGMGHD-LPEPLWNHIITKLTENFARSEPAAETPESEAAGR
MMAR_2912|M.marinum_M LKGAGHMYFHRRIWQQILSVLTEALA----------------
:.* ** : . :*: : * :