For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KPVKAPTGFDFFAHPELPGPPVSERDAEKFVVENYEISACARSLGSQQDANFLMTDPHGNTVGVLKVANA VFSTEEIDAQVEAVEWVAERDSALRLPRAIPDRRGQRHAPMSIGQEMLGVARLLTFLPGGAMSQSGYLAP KILAGMGNVTGRVSRALASFEHPGLDRVLQWDPRFASESVDALIGHVAEPAEAHRLAVATATAVNSLQRL DPQLPQQAVHLDIADTNMVGSPGSDGRRHPDGVIDFGDLTKTWAIAELATTVASCLYHAGAEPATILPVV AAFHAIRPLSGDEIEALWPMVIARAAVLLVSDLQQIAIDPGNEYAANTVQRERVILDQALLVPAPVMTGL IRDRLGIAQPQHHPLPKPHRPIIDIDPKSIVRLDFSVESDAANRGAWLDGALAHELADSALSSNALAAAT RYAATRADYIAPLHPRSSPTIATGIDFWLREDTELVAPWAGDVVASTGAITLRTDSHELQLTGAQPAPTS GQRVQAGQPWAKAPFGQRIHLALRPTDAPTAPHTVRPEYAPGWLAVTEDPAALLGLEPAPSDGLDVLERR AATFAEVQEHYYDDPPQIERGWRHYLLSTAGRSYLDMVNNVTVLGHAHPGVAAAAARQLERLNTNSRFNY AVVVEFCERLTQLLPDPLDTVFLVNSGSEAGDLALRLAMAATGRRDVVAVGEAYHGWTYATDAVSTSTAD NPNALNTRPDWVHTVESPNSFRGKYRGAGASQYAHDAVAAIEDLVQSGRAPAAFIAETVYGSAGGMALPD GYLDAVYTAIRAAGGLTIADEVQVGYGRLGKWFWGFEQQGVVPDIVAIAKATGNGHPVGAVITSKAIAQR FRSQGYFFSSTGGSPLSSAIGIAVLDALKSERLQHYALAVGSHLIARIRKLATKHPLIGTVHGFGLYIGV EMVRDRKTLEPAAEETAAICDRMLELGVIIQPTGERMNILKTKPPLCIDMTAADYFADTLDRVLTEGW*
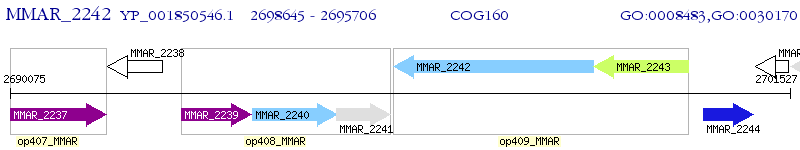
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2242 | gabT_1 | - | 100% (979) | 4-aminobutyrate aminotransferase GabT_1 |
| M. marinum M | MMAR_2465 | argD | 4e-35 | 29.15% (422) | acetylornithine aminotransferase ArgD |
| M. marinum M | MMAR_3558 | - | 3e-32 | 27.61% (431) | aminotransferase (adenosylmethionine-8-amino-7- oxononanoate) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1683 | argD | 1e-35 | 30.43% (414) | acetylornithine aminotransferase |
| M. gilvum PYR-GCK | Mflv_2095 | - | 0.0 | 57.76% (973) | hypothetical protein Mflv_2095 |
| M. tuberculosis H37Rv | Rv1655 | argD | 1e-35 | 30.43% (414) | acetylornithine aminotransferase |
| M. leprae Br4923 | MLBr_01409 | argD | 9e-36 | 30.02% (413) | acetylornithine aminotransferase |
| M. abscessus ATCC 19977 | MAB_0949c | - | 2e-73 | 38.02% (405) | aminotransferase |
| M. avium 104 | MAV_3115 | argD | 3e-35 | 30.00% (440) | acetylornithine aminotransferase |
| M. smegmatis MC2 155 | MSMEG_5211 | - | 0.0 | 56.06% (990) | hypothetical protein MSMEG_5211 |
| M. thermoresistible (build 8) | TH_3529 | - | 8e-35 | 30.58% (412) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_1647 | argD | 3e-34 | 29.38% (422) | acetylornithine aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_4610 | - | 0.0 | 58.61% (981) | hypothetical protein Mvan_4610 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK -----MSAGFNFLEQRELPAPAVDEVQAQRILAQSYGLDARVTSLGSQQD
Mvan_4610|M.vanbaalenii_PYR-1 -MSTRSTAGFNFLEQQELPAPQVSEAQAQDILATHYGLAAHVSALGSQQD
MSMEG_5211|M.smegmatis_MC2_155 MSAERSTVGFNFLAEPELPAPQVSEAQAEQILATHYGLQAKAASLGSQQD
MMAR_2242|M.marinum_M MKPVKAPTGFDFFAHPELPGPPVSERDAEKFVVENYEISACARSLGSQQD
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK KNFTVHGDDGTVLGVLKVANPAFTEVELCAQDEAARLIADTEPGLRVSVP
Mvan_4610|M.vanbaalenii_PYR-1 KNFTVRDDGGAVVGVLKIANPAFTAEELAAQDAAARRIAEAEPGLRVAVP
MSMEG_5211|M.smegmatis_MC2_155 KNFLVTQ-GDETVGVLKIANPAFNETELEAQDLAAELIAAAEPTLRIAVP
MMAR_2242|M.marinum_M ANFLMTDPHGNTVGVLKVANAVFSTEEIDAQVEAVEWVAERDSALRLPRA
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK LPNSAGEMYTTATDLVDG---TAFVRLLRYLPGGTLLESGYLPPAAVAGL
Mvan_4610|M.vanbaalenii_PYR-1 LANAAGETRTAVDGVLEG---TALVRLLQFLPGGTVSESGYLTPDSVAGL
MSMEG_5211|M.smegmatis_MC2_155 IPNVRGEKRTAVTGLLESRQATAYVRLLRFLPGDALMQRGYLRPSTVAEL
MMAR_2242|M.marinum_M IPDRRGQRHAPMSIGQEM---LGVARLLTFLPGGAMSQSGYLAPKILAGM
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK GDVAGRVSRALRDFTHPGLDRILQWDLRFGMDVVDALGPHVTDPALRSRL
Mvan_4610|M.vanbaalenii_PYR-1 GDVAGRVSRALADFTHPGLDRILQWDLRFGMHVVDELSAHVGEPALRRRL
MSMEG_5211|M.smegmatis_MC2_155 GALAGRVTRALAEFSHPGLDRILQWDLRYGAEVVATLASHVDDPARRDRL
MMAR_2242|M.marinum_M GNVTGRVSRALASFEHPGLDRVLQWDPRFASESVDALIGHVAEPAEAHRL
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK QEAARDAWSRLSALDDALPRQAVHLDLTDANVVVTRTPGAVS-PDGVIDF
Mvan_4610|M.vanbaalenii_PYR-1 QTAARDAWARIAPLDDALPRQAAHIDLTDANVVVS--PADGR-PDGVIDF
MSMEG_5211|M.smegmatis_MC2_155 EATTRDAWARIEPFADSLPHQAIHMDLTDANVVVSHEPGGVVRPDGIIDF
MMAR_2242|M.marinum_M AVATATAVNSLQRLDPQLPQQAVHLDIADTNMVGSPGSDGRRHPDGVIDF
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK GDLSHTWAVSELAITASSVLGHVGAEVTSTLPAIGAFHTVRPLSDAEVGA
Mvan_4610|M.vanbaalenii_PYR-1 GDLSHTWAVSELAITASSVLGHVGAQVTSVLPAIRAFHAVRPLSVAEADA
MSMEG_5211|M.smegmatis_MC2_155 GDLTDSWTVGELAITLSSVLQHPGATPTSVLPGIRAFHAVRPLSVDEAMV
MMAR_2242|M.marinum_M GDLTKTWAIAELATTVASCLYHAGAEPATILPVVAAFHAIRPLSGDEIEA
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK LWPMVVLRTVVLIVSDAQQAVLDPDNDYIAAQSDSGMRMFEAATSVPIDV
Mvan_4610|M.vanbaalenii_PYR-1 LWPMLVLRTAVLIVSGAQQSVLDPDNEYLTEQSDAEQQMFDLATSVPIDV
MSMEG_5211|M.smegmatis_MC2_155 LWPLLVLRTAVLNVSGAQQAAIDPDNAYVTENADGEWQMFEAATSVPLDV
MMAR_2242|M.marinum_M LWPMVIARAAVLLVSDLQQIAIDPGNEYAANTVQRERVILDQALLVPAPV
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK MTEVIAAYLGQGQPR--PPVRPAALMIA-VDRATAVTLDLSTTSDLYDDG
Mvan_4610|M.vanbaalenii_PYR-1 MTAVIKAGLGMAQPS--PPVRVQAQLIG-ADRTSTVTLDLSTTSEVYDDA
MSMEG_5211|M.smegmatis_MC2_155 MTALITADLGFDCAA--PELGALTPMITGLEADAVTTLDLGTVSDTLDSA
MMAR_2242|M.marinum_M MTGLIRDRLGIAQPQHHPLPKPHRPIID-IDPKSIVRLDFSVESDAANRG
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK FATAG--------DHAGAALRRGAPVVVTRHGEPRLCNAPRLSQEAPDVV
Mvan_4610|M.vanbaalenii_PYR-1 FDAAGVMRSDIEDESARAAMHQGATVVVTRFGEARLDRAPRLSQDSPEVV
MSMEG_5211|M.smegmatis_MC2_155 FTRGGWVRSDIENELARAALADGARVVLTQFGQPRPTRAPALSHDEPAVV
MMAR_2242|M.marinum_M AWLDG----ALAHELADSALSSNALAAATRYAATRADYIAPLHPRSSPTI
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK ATGIELWTAARTELAAPWDGEISEEVSGRLTLRGNEFELTLAGVTEP--V
Mvan_4610|M.vanbaalenii_PYR-1 ATGISMWTAADTDIAAPWDGEVVTDATGSITLRGNDFEVTVVGAAPA--G
MSMEG_5211|M.smegmatis_MC2_155 PTGITMWCAEDVALTAPFEGEISRDGD-SVTLRGNTCELTLTGVSG----
MMAR_2242|M.marinum_M ATGIDFWLREDTELVAPWAGDVVASTG-AITLRTDSHELQLTGAQPAPTS
MAB_0949c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3115|M.avium_104 --------------------------------------------------
MUL_1647|M.ulcerans_Agy99 --------------------------------------------------
Mb1683|M.bovis_AF2122/97 --------------------------------------------------
Rv1655|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01409|M.leprae_Br4923 --------------------------------------------------
TH_3529|M.thermoresistible__bu --------------------------------------------------
Mflv_2095|M.gilvum_PYR-GCK RGTVSAGDVIGSAAQHGPVEISVRPVGAPVAPMFVRSDLAAGWLAVTRDP
Mvan_4610|M.vanbaalenii_PYR-1 GAAVCAGEVLASARAGERIEVSVRPVGVPVAPPFIRADLAPGWLAQVRDP
MSMEG_5211|M.smegmatis_MC2_155 -----DADTFGVARAGRPVEVALRPVGAPVAPALVTPSLAPGWLAWSVDP
MMAR_2242|M.marinum_M GQRVQAGQPWAKAPFGQRIHLALRPTDAPTAPHTVRPEYAPGWLAVTEDP
MAB_0949c|M.abscessus_ATCC_199 -----------------------------------------MSFSNIMDS
MAV_3115|M.avium_104 -------------MVTNTDALRRRWEAVMMHNYGTPPVALASGEGAVVTD
MUL_1647|M.ulcerans_Agy99 --------------MTHTEDTRARWEAVMMNNYGTPPVALASGEGAVVTD
Mb1683|M.bovis_AF2122/97 ----------MTGASTTTATMRQRWQAVMMNNYGTPPIALASGDGAVVTD
Rv1655|M.tuberculosis_H37Rv ----------MTGASTTTATMRQRWQAVMMNNYGTPPIALASGDGAVVTD
MLBr_01409|M.leprae_Br4923 ----------MTPTQTNTATMQQRWETVMMNNYGTPPIVLASGNGAVVTD
TH_3529|M.thermoresistible__bu -----------VSGSKGRHTLLDRWQGVMMNNYGTPPLALVSGDGAVVTD
Mflv_2095|M.gilvum_PYR-GCK RPLLGLPPLTTDDR-DDLLARRDASFAPVQEFYYRTPPQIERGRRHYLMS
Mvan_4610|M.vanbaalenii_PYR-1 RPLLGLAPLEQDGA-ADLLSRRDASFAPVQEFYYRTPPQIERGRRHYLMS
MSMEG_5211|M.smegmatis_MC2_155 RPLFGLPALATDSTDADLVARRDRAFAPVQEHYYDEPPRIERGWRHFLMS
MMAR_2242|M.marinum_M AALLGLEPAPSDGL--DVLERRAATFAEVQEHYYDDPPQIERGWRHYLLS
MAB_0949c|M.abscessus_ATCC_199 NSYAGVHDDPATES---LITARERVLGPSYRLFYERPVHLVRGQGSHLFD
. : * : * : .
MAV_3115|M.avium_104 VDGNTYLDLLGGIAVNVLGHRHPAIIEAVTKQLSTLGHTSNLYATEPSLA
MUL_1647|M.ulcerans_Agy99 VDGKNYVDLLGGIAVNILGHRHPAVIEAVTQQMSTLGHTSNLYATEPTNA
Mb1683|M.bovis_AF2122/97 VDGRTYIDLLGGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEPGIA
Rv1655|M.tuberculosis_H37Rv VDGRTYIDLLGGIAVNVLGHRHPAVIEAVTRQMSTLGHTSNLYATEPGIA
MLBr_01409|M.leprae_Br4923 VDSNTYLDLLGGIAVNVLGHRHPAVIEAVTHQITTLGHTSNLYATEPSIT
TH_3529|M.thermoresistible__bu ADGKSYLDLLGGIAVNILGHRHPAVIEAVTRQLNTLGHTSNLYATEPGVA
Mflv_2095|M.gilvum_PYR-GCK TRARTYLDMVN--NVTVLGHAHPRIAEVAARQLHRLNTNS-RFNYEAVVA
Mvan_4610|M.vanbaalenii_PYR-1 TAGRSYLDMVN--NVTVLGHAHPRIADTAARQLRRLNTNS-RFNYEAVVE
MSMEG_5211|M.smegmatis_MC2_155 TSGRCYLDMVN--NVTVLGHAHPRIAEAAARQLRKLNTNS-RFNYASVVE
MMAR_2242|M.marinum_M TAGRSYLDMVN--NVTVLGHAHPGVAAAAARQLERLNTNS-RFNYAVVVE
MAB_0949c|M.abscessus_ATCC_199 NEGNRYLDAYN--NVASVGHCHPHVVGAVTRQLSALNTHT-RYLHEGIVD
.. *:* . * :** ** : ..::*: *. : :
MAV_3115|M.avium_104 LAEELVALLGAGHEPAGHESGGTESAGTARVFFCNSGTEANEVAFKLS-R
MUL_1647|M.ulcerans_Agy99 LAEELVTLLGA--------------EAPARVFFCNSGTEANEAAFKLS-R
Mb1683|M.bovis_AF2122/97 LAEELVALLGA--------------DQRTRVFFCNSGAEANEAAFKLS-R
Rv1655|M.tuberculosis_H37Rv LAEELVALLGA--------------DQRTRVFFCNSGAEANEAAFKLS-R
MLBr_01409|M.leprae_Br4923 LAEELVALLGA--------------DTQTRVFFCNSGTEANELAFKLS-R
TH_3529|M.thermoresistible__bu LAEALVGHLGA----------------PARVFFCNSGAEANEVAFKIS-R
Mflv_2095|M.gilvum_PYR-GCK FSERLAGLLPD---------------PLDTVFLVNSGSEASDLAIRLATA
Mvan_4610|M.vanbaalenii_PYR-1 FSERLAALLPD---------------PLDTVFLVNSGSEASDLAIRLATA
MSMEG_5211|M.smegmatis_MC2_155 FSERLASLLPD---------------PLDTVFLVNSGSEASDLALRLAIA
MMAR_2242|M.marinum_M FCERLTQLLPD---------------PLDTVFLVNSGSEAGDLALRLAMA
MAB_0949c|M.abscessus_ATCC_199 YSERLLATMPT---------------PIDQVMYACTGSEANDLALRVAQR
.* * : *: :*:**.: *::::
MAV_3115|M.avium_104 LTGRTKLVAAQEAFHGRTMGSLALTGQPSKQAPFAPLPGDVTHVPYG---
MUL_1647|M.ulcerans_Agy99 LTGRTKLVAAQEGFHGRTMGSLALTGQPAKQAPFEPLPGYVTHVPYG---
Mb1683|M.bovis_AF2122/97 LTGRTKLVAAHDAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYG---
Rv1655|M.tuberculosis_H37Rv LTGRTKLVAAHDAFHGRTMGSLALTGQPAKQTPFAPLPGDVTHVGYG---
MLBr_01409|M.leprae_Br4923 LTGRTKLVAAQAAFHGRTMGSLALTGQPAKQAAFEPLPGHVTHVPYG---
TH_3529|M.thermoresistible__bu LTGRTKVVAAEGGFHGRTMGALALTGQPSKREPFEPLPGDVIHVPYG---
Mflv_2095|M.gilvum_PYR-GCK ATGRRDVVAVREAYHGWTYGTDAVSTSTADNPNALATRPDWVHTVESPNS
Mvan_4610|M.vanbaalenii_PYR-1 ATGRRDVVAVREAYHGWTYGTDAVSTSTADNPNALATRPDWVHTVESPNS
MSMEG_5211|M.smegmatis_MC2_155 AAGRPDVVAMREAYHGWTYGTDAVSTSTADNPNALETRPDWVHTVESPNS
MMAR_2242|M.marinum_M ATGRRDVVAVGEAYHGWTYATDAVSTSTADNPNALNTRPDWVHTVESPNS
MAB_0949c|M.abscessus_ATCC_199 YTGHKGIVVTSDAYHGNTAAVTAISPSLGGN-----SLGAHVRTVAPPDS
:*: :*. .:** * . *:: . . . :.
MAV_3115|M.avium_104 ------------------DTEALAAAVDDGTAAVFLEPIMGESGVVVPPE
MUL_1647|M.ulcerans_Agy99 ------------------DIDALAAAVDDETAAVFLEPIMGESGVVVPPE
Mb1683|M.bovis_AF2122/97 ------------------DVDALAAAVDDHTAAVFLEPIMGESGVVVPPA
Rv1655|M.tuberculosis_H37Rv ------------------DVDALAAAVDDHTAAVFLEPIMGESGVVVPPA
MLBr_01409|M.leprae_Br4923 ------------------QVDALAAAVDNDTAAVFLEPIMGESGVIVPPE
TH_3529|M.thermoresistible__bu ------------------DAGALERAVDDTTAAVFLEPIMGEGGVVVPPA
Mflv_2095|M.gilvum_PYR-GCK FRGMYRGAEASRYAVDAVAQIEALAASGRPPAAFICESVYGNAGGMALPD
Mvan_4610|M.vanbaalenii_PYR-1 FRGKYRGSEAFRYAEDAVAQIEALVMSGRPPAAFICESVYGNAGGMALPD
MSMEG_5211|M.smegmatis_MC2_155 FRGKYRGSDVGRYADEAVAQIQELAAAGRAPAAFICETVYGNAGGMALPD
MMAR_2242|M.marinum_M FRGKYRGAGASQYAHDAVAAIEDLVQSGRAPAAFIAETVYGSAGGMALPD
MAB_0949c|M.abscessus_ATCC_199 YR-TPPGELAERFAADVSHAIDELCAAGYGFSCLVVDTIFSSDG-IYPDA
. :... :.: .. * :
MAV_3115|M.avium_104 GYLAAARDITARHGALLVLDEVQTGMGRTG-TFFAHQHDGITPDVVTLAK
MUL_1647|M.ulcerans_Agy99 GYLLAAREITARHGALLVLDEVQTGIGRTG-AFYAHQHDGITPDVVTLAK
Mb1683|M.bovis_AF2122/97 GYLAAARDITARRGALLVLDEVQTGMGRTG-AFFAHQHDGITPDVVTLAK
Rv1655|M.tuberculosis_H37Rv GYLAAARDITARRGALLVLDEVQTGMGRTG-AFFAHQHDGITPDVVTLAK
MLBr_01409|M.leprae_Br4923 GYLAAARDITTRHGALLVIDEVQTGIGRTG-AFFAHQHDSITPDVVTLAK
TH_3529|M.thermoresistible__bu GYLAAAREVTARHGALLVLDEVQTGIGRTG-TFFAHQHDGITPDVVTLAK
Mflv_2095|M.gilvum_PYR-GCK GYLRQIYAAIRAGGGLAISDEVQVGYGRLGQWFWGFQQQDAVPDIVSVAK
Mvan_4610|M.vanbaalenii_PYR-1 GYLKQVYAAVRAGGGLAISDEVQVGYGRLGEWFWGFQQQDAVPDIVSVAK
MSMEG_5211|M.smegmatis_MC2_155 GYLEKVYAAIRANGGYAVADEVQVGYGRLGEWFWGFQQQGVVPDIVSMAK
MMAR_2242|M.marinum_M GYLDAVYTAIRAAGGLTIADEVQVGYGRLGKWFWGFEQQGVVPDIVAIAK
MAB_0949c|M.abscessus_ATCC_199 SVLEPAAEVVRGAGGVLIADEVQPGFGRTGETWWGFIRHGVVPDLVTMGK
. * *. : **** * ** * ::.. :.. .**:*::.*
MAV_3115|M.avium_104 GLGGGLPIGACLAVGPAADLLTP-GLHGSTFGGNPVCAAAALAVLRVLAS
MUL_1647|M.ulcerans_Agy99 GLGGGLPIGACLAIGPAAELLTP-GLHGSTFGGNPVCAAAALAVLRVVAS
Mb1683|M.bovis_AF2122/97 GLGGGLPIGACLAVGPAAELLTP-GLHGSTFGGNPVCAAAALAVLRVLAS
Rv1655|M.tuberculosis_H37Rv GLGGGLPIGACLAVGPAAELLTP-GLHGSTFGGNPVCAAAALAVLRVLAS
MLBr_01409|M.leprae_Br4923 GLGGGLPIGAFLATGPAAELLTL-GLHGSTFGGNPVCTAAALAVLRVLAT
TH_3529|M.thermoresistible__bu GLGGGLPIGACLAVGPAGDLLTP-GMHGSTFGGNPVCTAAAVAVLGALAE
Mflv_2095|M.gilvum_PYR-GCK SVGNGYPVGAVITTRAVAQAFATEGYFFSSTGGSPLSCAIGMAVLDVLHE
Mvan_4610|M.vanbaalenii_PYR-1 SVGNGYPVGAVITTRAVAEAFSSQGYFFSSTGGSPLSCAIGMTVLDVLRD
MSMEG_5211|M.smegmatis_MC2_155 STGNGYPLGAVVTSRAVAEAFRSQGYFFSSTGGSPLSCALGMTVLDVLGE
MMAR_2242|M.marinum_M ATGNGHPVGAVITSKAIAQRFRSQGYFFSSTGGSPLSSAIGIAVLDALKS
MAB_0949c|M.abscessus_ATCC_199 PMANGIPVSALAATSEVLAPFAQDVPYFNTFGGNPVSMAAAAAVMDVIED
..* *:.* : : . .: **.*:. * . :*: .:
MAV_3115|M.avium_104 ENLVRHAEVLGKSLHQGIEGLG--HPLVDHVRGRGLLCGVVLTAPRAKDA
MUL_1647|M.ulcerans_Agy99 EGLVCRAETLGKTFKQGVESLS--HPLIDHVRGRGLLLGIVLAGPHAKAA
Mb1683|M.bovis_AF2122/97 DGLVRRAEVLGKSLRHGIEALG--HPLIDHVRGRGLLLGIALTAPHAKDA
Rv1655|M.tuberculosis_H37Rv DGLVRRAEVLGKSLRHGIEALG--HPLIDHVRGRGLLLGIALTAPHAKDA
MLBr_01409|M.leprae_Br4923 QGLVRRAEVLGDSMRIGIESLS--HPLIDQVRGRGLLLGIVLTAPRAKDI
TH_3529|M.thermoresistible__bu GDLINRADVLGKKLSHGIEELR--HPLVDHVRGRGLLCGVVLNAPQAKAA
Mflv_2095|M.gilvum_PYR-GCK EGLQDNAFRVGGHLKGRLEGLRDKHPLIGTVHGFGLYLGVEMVRDPQTLE
Mvan_4610|M.vanbaalenii_PYR-1 EGLQDNARRVGTHLKTRLEGLKERHPLVGTVHGFGLYLGVEMIRDPQTLT
MSMEG_5211|M.smegmatis_MC2_155 ERLQENASRVGAHLKSRLLALSDKHPIIGTVHGVGLYLGVEMIRDPATLE
MMAR_2242|M.marinum_M ERLQHYALAVGSHLIARIRKLATKHPLIGTVHGFGLYIGVEMVRDRKTLE
MAB_0949c|M.abscessus_ATCC_199 EKLQLHAAEVGAMLRTELRSLASTHVRLGDVRGTGLYTGVEVVDDPAART
* * :* : : * * :. *:* ** *: :
MAV_3115|M.avium_104 EAAAR---------EAGFLVNAAAP--GVIRLAPPLIVTEAQIDGFIAAL
MUL_1647|M.ulcerans_Agy99 EVAAR---------EVGYLVNGAAP--NVVRLAPPLVITEPQLEGFVAAL
Mb1683|M.bovis_AF2122/97 EATAR---------DAGYLVNAAAP--DVIRLAPPLIIAEAQLDGFVAAL
Rv1655|M.tuberculosis_H37Rv EATAR---------DAGYLVNAAAP--DVIRLAPPLIIAEAQLDGFVAAL
MLBr_01409|M.leprae_Br4923 EKAAR---------DAGFLVNATAP--EVIRLAPPLIITESQIDSFITAL
TH_3529|M.thermoresistible__bu EAAAR---------DAGFLVNAAAP--EVIRLAPPLIITETQIDEFLAAL
Mflv_2095|M.gilvum_PYR-GCK PAPEETSAICDRMLDLGVVIQPTGDHQNILKTKPPLCIDVEAADFYVDTL
Mvan_4610|M.vanbaalenii_PYR-1 PATAETSAICDRMLDLGVIIQPTGDHQNILKTKPPLCIDVEAADFYVDTL
MSMEG_5211|M.smegmatis_MC2_155 PATEETALICDRMLELGVIIQPTGDHQNILKTKPPLCIDTESADFYVDTL
MMAR_2242|M.marinum_M PAAEETAAICDRMLELGVIIQPTGERMNILKTKPPLCIDMTAADYFADTL
MAB_0949c|M.abscessus_ATCC_199 PDRAGALHIVNELRNRRVLISVCGQDGNVLKIRPPLVFSRSDVDWFCTEL
: ::. . ::: *** . : : *
MAV_3115|M.avium_104 PGILDRAAA------
MUL_1647|M.ulcerans_Agy99 PGILDNAGASA----
Mb1683|M.bovis_AF2122/97 PAILDRAVGAP----
Rv1655|M.tuberculosis_H37Rv PAILDRAVGAP----
MLBr_01409|M.leprae_Br4923 PGILDASIAELGKKA
TH_3529|M.thermoresistible__bu PGVLDTAKEAS----
Mflv_2095|M.gilvum_PYR-GCK DRVLTEGW-------
Mvan_4610|M.vanbaalenii_PYR-1 DRVLTEGW-------
MSMEG_5211|M.smegmatis_MC2_155 DRVLTEGW-------
MMAR_2242|M.marinum_M DRVLTEGW-------
MAB_0949c|M.abscessus_ATCC_199 DDVLKSTG-------
:*