For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QALTIAALSRAQVDIPIDQLPSLPPLSDELRARLDAALAKPAAQQPSWPVDQATAMRKVLEGVPPVTVPS EIIRLQEQLAQVARGEAFLLQGGDCAETFTDNTEPHIRGNVRTLLQMAVVLTYGSSMPVVKVARIAGQYA KPRSADTDALGLKSYRGDMINGFPPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDWNR EFVRTSPAGARYEALANEIDRGLRFMSACGVADRNLQTAEIYASHEALVLDYERAMLRLGEGENGEPQLY DLSAHTVWIGERTRQLDGAHVAFAEVIANPIGVKIGPTMTPELAVEYVERLDPHNKAGRLTLVSRLGNNK VRDLLPPIVEKVQAAGHQVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGFFEVHRGLGTHPGGIHVEI TGENVTECLGGAQDISDSDLSGRYETACDPRLNTQQSLELAFLVAEMLRN*
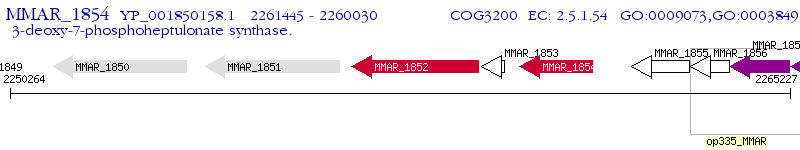
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1854 | aroG_1 | - | 100% (471) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG_1 |
| M. marinum M | MMAR_3222 | aroG | 0.0 | 99.56% (458) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2200c | aroG | 0.0 | 91.70% (458) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. gilvum PYR-GCK | Mflv_2971 | - | 0.0 | 86.21% (464) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. tuberculosis H37Rv | Rv2178c | aroG | 0.0 | 91.48% (458) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. leprae Br4923 | MLBr_00896 | aroG | 0.0 | 90.24% (461) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAHP |
| M. abscessus ATCC 19977 | MAB_1987 | - | 0.0 | 86.68% (458) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. avium 104 | MAV_2317 | - | 0.0 | 92.79% (458) | 3-deoxy-7-phosphoheptulonate synthase |
| M. smegmatis MC2 155 | MSMEG_4244 | - | 0.0 | 87.83% (460) | 3-deoxy-7-phosphoheptulonate synthase |
| M. thermoresistible (build 8) | TH_1102 | aroG | 0.0 | 88.21% (458) | Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate |
| M. ulcerans Agy99 | MUL_3533 | aroG_1 | 0.0 | 99.13% (458) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_3540 | - | 0.0 | 86.18% (463) | phospho-2-dehydro-3-deoxyheptonate aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2971|M.gilvum_PYR-GCK ---------MNWTVDVPIDQLPALPPLPADLRQRLDAALAKPALQQPSWD
Mvan_3540|M.vanbaalenii_PYR-1 ---------MNWTVDVPIDQLPALPPLPEDLRQRLDAALARPALQQPSWD
MSMEG_4244|M.smegmatis_MC2_155 ---------MNWTVDIPIDQLPPLPPLSDELRQRLDSALAKPAVQQPSWD
TH_1102|M.thermoresistible__bu ---------VNWTVDVPIDQLPALPPLPADLRQRLDAALAKPALQQPSWD
MMAR_1854|M.marinum_M MQALTIAALSRAQVDIPIDQLPSLPPLSDELRARLDAALAKPAAQQPSWP
MUL_3533|M.ulcerans_Agy99 ---------MNWTVDIPIDQLPSLPPLSDELRARLDAALAKPAAQQPSWP
Mb2200c|M.bovis_AF2122/97 ---------MNWTVDIPIDQLPSLPPLPTDLRTRLDAALAKPAAQQPTWP
Rv2178c|M.tuberculosis_H37Rv ---------MNWTVDIPIDQLPSLPPLPTDLRTRLDAALAKPAAQQPTWP
MAV_2317|M.avium_104 ---------MNWTVDIPIDQLPPLPPLPGDLRARLDAALAKPAAQQPSWP
MLBr_00896|M.leprae_Br4923 ---------MNWTVDIPIDQLP---PLPADLRARLDAALAKPAAQQPSWP
MAB_1987|M.abscessus_ATCC_1997 ---------MNWTVDVPIDQLPELPPLPADLRERLDAALAKPAAQQPSWP
. **:****** **. :** ***:***:** ***:*
Mflv_2971|M.gilvum_PYR-GCK AGQAAAMRKVLESVPPVTVPSEIERLKGQLADVALGKAFLLQGGDCAETF
Mvan_3540|M.vanbaalenii_PYR-1 AGQAAAMRKVLESVPPVTVPSEIERLKSQLADVARGQAFLLQGGDCAETF
MSMEG_4244|M.smegmatis_MC2_155 PDAAKAMRTVLESVPPVTVPSEIEKLKGLLADVAQGKAFLLQGGDCAETF
TH_1102|M.thermoresistible__bu PDLAKAMRTVLESVPPITVPSEVERLKSLLGEVARGKAFLLQGGDCAETF
MMAR_1854|M.marinum_M VDQATAMRKVLEGVPPVTVPSEIIRLQEQLAQVARGEAFLLQGGDCAETF
MUL_3533|M.ulcerans_Agy99 VDQATAMRKVLESVPPVTVPSEIIRLQEQLAQVAGGEAFLLQGGDCAETF
Mb2200c|M.bovis_AF2122/97 ADQALAMRTVLESVPPVTVPSEIVRLQEQLAQVAKGEAFLLQGGDCAETF
Rv2178c|M.tuberculosis_H37Rv ADQALAMRTVLESVPPVTVPSEIVRLQEQLAQVAKGEAFLLQGGDCAETF
MAV_2317|M.avium_104 TDQATAMRTVLESVPPVTVPSEIVRLQEQLAQVARGEAFLLQGGDCAETF
MLBr_00896|M.leprae_Br4923 TDQAAAMRTVLESVPPVTVPSEIIRLQEQLALVANGKAFLLQGGDCAETF
MAB_1987|M.abscessus_ATCC_1997 ANQAAAMRTVLESVPPITVPAEIQRLQRQLAQVARGEAFLLQGGDCAETF
. * ***.***.***:***:*: :*: *. ** *:*************
Mflv_2971|M.gilvum_PYR-GCK VDNTEPHIRANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDVDA
Mvan_3540|M.vanbaalenii_PYR-1 VDNTEPHIRANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDTDA
MSMEG_4244|M.smegmatis_MC2_155 VDNTEPHIRANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDVDA
TH_1102|M.thermoresistible__bu VDNTEPHIKANIRTLLQMAVVLTYGASTPVVKMARIAGQYAKPRSSDTDA
MMAR_1854|M.marinum_M TDNTEPHIRGNVRTLLQMAVVLTYGSSMPVVKVARIAGQYAKPRSADTDA
MUL_3533|M.ulcerans_Agy99 TDNTEPHIRGNVRTLLQMAVVLTYGSSMPVVKVARIAGQYAKPRSADTDA
Mb2200c|M.bovis_AF2122/97 MDNTEPHIRGNVRALLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDA
Rv2178c|M.tuberculosis_H37Rv MDNTEPHIRGNVRALLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDA
MAV_2317|M.avium_104 TENTEPHIRGNIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDA
MLBr_00896|M.leprae_Br4923 VDNTEPHIRSNVRTLLQMAVVLTYGASLPVVKVARIAGQYAKPRSADVDA
MAB_1987|M.abscessus_ATCC_1997 ADNTEPHIRANIRALLQMAVVLTYGASMPVVKLARIAGQYAKPRSSDTDA
:******:.*:*:***********:* ****:************:* **
Mflv_2971|M.gilvum_PYR-GCK LGLKSYRGDMVNGFAPDAAVRDHDPSRLVRAYANASAAMNLVRALTSSGM
Mvan_3540|M.vanbaalenii_PYR-1 LGLKSYRGDMVNGFAPDAAVREHDPSRLVRAYANASAAMNLVRALTSSGM
MSMEG_4244|M.smegmatis_MC2_155 LGLKSYRGDMINGFAPDAAAREHDPSRLVRAYANASAAMNLMRALTSSGL
TH_1102|M.thermoresistible__bu LGLKSYRGDMINGFAPDASAREHDPSRLVRAYANASAAMNLVRALTSSGL
MMAR_1854|M.marinum_M LGLKSYRGDMINGFPPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGL
MUL_3533|M.ulcerans_Agy99 LGLKSYRGDMINGFPPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGL
Mb2200c|M.bovis_AF2122/97 LGLRSYRGDMINGFAPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGL
Rv2178c|M.tuberculosis_H37Rv LGLRSYRGDMINGFAPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGL
MAV_2317|M.avium_104 LGLKSYRGDMINGFAPNAAVREHDPSRLVRAYANASAAMNLVRALTSSGL
MLBr_00896|M.leprae_Br4923 LGLKSYRGDMINGFAPDAAAREHDPSRLVRAYANSSAAMNLVRALTSSGL
MAB_1987|M.abscessus_ATCC_1997 LGLKSYRGDMVNGFAPDATLREHDPSRLVRAYANASAAMNLVRALTGSGM
***:******:***.*:*: *:************:******:****.**:
Mflv_2971|M.gilvum_PYR-GCK ASLHQVHDWNREFVRTSPAGARYEALAGEIDRGLRFMSACRVDDRNLDTA
Mvan_3540|M.vanbaalenii_PYR-1 ASLHQVHDWNREFVRTSPAGARYEALAGEIDRGLRFMSACRVDDRNLDTA
MSMEG_4244|M.smegmatis_MC2_155 ASLHLVHEWNREFVRTSPAGARYEALAGEIDRGLNFMSACGVADRNLQTA
TH_1102|M.thermoresistible__bu ASLHLVHDWNREFVRTSPAGARYEALAGEIDRGLRFMSACGVADRNLQTA
MMAR_1854|M.marinum_M ASLHLVHDWNREFVRTSPAGARYEALANEIDRGLRFMSACGVADRNLQTA
MUL_3533|M.ulcerans_Agy99 ASLHLVHDWNREFVRTSPAGARYEALANEIDRGLRFMSACGVADRNLQTA
Mb2200c|M.bovis_AF2122/97 ASLHLVHDWNREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTA
Rv2178c|M.tuberculosis_H37Rv ASLHLVHDWNREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTA
MAV_2317|M.avium_104 ASLHLVHDWNREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTA
MLBr_00896|M.leprae_Br4923 ASLHLVHDWNREFVRTSPAGARYEALAGEIGRGLAFMSACGVADRNLQTA
MAB_1987|M.abscessus_ATCC_1997 ASLALVHDWNREFVRTSPAGARYEALAGEIDRGLKFMSACGVVDSNLDTA
*** **:******************* **.*** ***** * * **:**
Mflv_2971|M.gilvum_PYR-GCK EIYASHEALVLDYERAMLRMETGDLADPALSSEPKLYDLSAHYVWIGERT
Mvan_3540|M.vanbaalenii_PYR-1 EIYASHEALVLDYERAMLRMNIADSADPASDGPPKLYDLSAHYVWIGERT
MSMEG_4244|M.smegmatis_MC2_155 EIFASHEALVLDYERAMLRLSN----PAETDGAAKLYDQSAHYLWIGERT
TH_1102|M.thermoresistible__bu EIYASHEALVLDYERAMLRLS-------EETGEPKLYDLSAHYLWIGERT
MMAR_1854|M.marinum_M EIYASHEALVLDYERAMLRLG------EGENGEPQLYDLSAHTVWIGERT
MUL_3533|M.ulcerans_Agy99 EIYASHEALVLDYERVMLRLG------EGENGEPQLYDLSAHTVWIGERT
Mb2200c|M.bovis_AF2122/97 EIYASHEALVLDYERAMLRLS------DGEDGEPQLFDLSAHTVWIGERT
Rv2178c|M.tuberculosis_H37Rv EIYASHEALVLDYERAMLRLS------DGDDGEPQLFDLSAHTVWIGERT
MAV_2317|M.avium_104 EIYASHEALVLDYERAMLRLS------DSDAGEPQLYDLSAHTVWIGERT
MLBr_00896|M.leprae_Br4923 EIYASHEALVLDYERAMLRLAG---APEGPDDGLQLYDLSAHTVWIGERT
MAB_1987|M.abscessus_ATCC_1997 EIYASHEALVLDYERAMLRLG------EDENGEPALYDLSAHYLWIGDRT
**:************.***: . *:* *** :***:**
Mflv_2971|M.gilvum_PYR-GCK RQLDGAHVAFAEVIANPIGIKIGPTTSPELAVEYVERLDPNNEPGRLTLV
Mvan_3540|M.vanbaalenii_PYR-1 RQLDGAHVAFAEVIANPVGIKIGPTTSPELAVEYVERLDPNNEPGRLTLV
MSMEG_4244|M.smegmatis_MC2_155 RQLDGAHVAFAEVIANPIGVKLGPTTTPELAVEYVERLDPNNEPGRLTLV
TH_1102|M.thermoresistible__bu RQLDGAHIAFAEVIANPIGVKIGPTTTPEVAAEYVERLDPNNEPGRLTLV
MMAR_1854|M.marinum_M RQLDGAHVAFAEVIANPIGVKIGPTMTPELAVEYVERLDPHNKAGRLTLV
MUL_3533|M.ulcerans_Agy99 RQLDGAHVAFAEVIANPIGVKIGPTMTPELAVEYVERLDPHNKAGRLTLV
Mb2200c|M.bovis_AF2122/97 RQIDGAHIAFAQVIANPVGVKLGPNMTPELAVEYVERLDPHNKPGRLTLV
Rv2178c|M.tuberculosis_H37Rv RQIDGAHIAFAQVIANPVGVKLGPNMTPELAVEYVERLDPHNKPGRLTLV
MAV_2317|M.avium_104 RQLDGAHIAFAEVIANPIGVKIGPTITPELAVEYVERLDPHNKPGRLTLV
MLBr_00896|M.leprae_Br4923 RQLDGAHVAFAEVIANPIGVKMGATMTPELAVEYVERLDPHNKPGRLTLV
MAB_1987|M.abscessus_ATCC_1997 RQLDHAHVAFAEIIANPIGMKIGPTTTPDQAVEYVERLDPHNKPGRLTFV
**:* **:***::****:*:*:*.. :*: *.********:*:.****:*
Mflv_2971|M.gilvum_PYR-GCK SRMGNHKVRDVLPPIIEKVQASGHRVIWQCDPMHGNTHESSTGYKTRHFD
Mvan_3540|M.vanbaalenii_PYR-1 SRMGNHKVRDVLPPIIEKVQASGHQVVWQCDPMHGNTHESSTGYKTRHFD
MSMEG_4244|M.smegmatis_MC2_155 TRMGNNKVRDLLPPIIEKVQATGHQVIWQCDPMHGNTHESSTGYKTRHFD
TH_1102|M.thermoresistible__bu SRLGNHKVRDLLPPIIEKVQATGHTVIWQCDPMHGNTHESSTGYKTRHFD
MMAR_1854|M.marinum_M SRLGNNKVRDLLPPIVEKVQAAGHQVIWQCDPMHGNTHESSTGYKTRHFD
MUL_3533|M.ulcerans_Agy99 SRLGNNKVRDLLPPIVEKVQAAGHQVIWQCDPMHGNTHESSTGYKTRHFD
Mb2200c|M.bovis_AF2122/97 SRMGNHKVRDLLPPIVEKVQATGHQVIWQCDPMHGNTHESSTGFKTRHFD
Rv2178c|M.tuberculosis_H37Rv SRMGNHKVRDLLPPIVEKVQATGHQVIWQCDPMHGNTHESSTGFKTRHFD
MAV_2317|M.avium_104 SRMGNSKVRDLLPPIVEKVQATGHQVIWQCDPMHGNTHESSNGYKTRHFD
MLBr_00896|M.leprae_Br4923 SRLGNNKVRDVLPPIVEKVKATGHQVIWQCDPMHGNTHESSTGYKTRHFD
MAB_1987|M.abscessus_ATCC_1997 SRMGNSKVRDLLPPIIEKVRATGHQVVWQCDPMHGNTHESPSGYKTRHFD
:*:** ****:****:***:*:** *:*************..*:******
Mflv_2971|M.gilvum_PYR-GCK RIVDEVQGFFEVHRALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGR
Mvan_3540|M.vanbaalenii_PYR-1 RIVDEVQGFFEVHHALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGR
MSMEG_4244|M.smegmatis_MC2_155 RIVDEVQGFFEVHHALGTHPGGIHVEITGENVTECLGGAQDISDSDLAGR
TH_1102|M.thermoresistible__bu RIVDEVQGFFEVHRALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGR
MMAR_1854|M.marinum_M RIVDEVQGFFEVHRGLGTHPGGIHVEITGENVTECLGGAQDISDSDLSGR
MUL_3533|M.ulcerans_Agy99 RIVDEVQGFFEVHRGLGTHPGGIHVEITGENVTECLGGAQDISDSDLSGR
Mb2200c|M.bovis_AF2122/97 RIVDEVQGFFEVHRALGTHPGGIHVEITGENVTECLGGAQDISETDLAGR
Rv2178c|M.tuberculosis_H37Rv RIVDEVQGFFEVHRALGTHPGGIHVEITGENVTECLGGAQDISETDLAGR
MAV_2317|M.avium_104 RIVDEVQGFFEVHRALGTHPGGIHVEITGDNVTECLGGAQDISDMDLVGR
MLBr_00896|M.leprae_Br4923 RVVDEVQGFFEVHRALGTYPGGIHVEITGEDVTECLGGAQDISDTDLAGR
MAB_1987|M.abscessus_ATCC_1997 RIVDEVQGYFEVHNALGTHPGGIHVEITGENVTECLGGAQDISDDDLAGR
*:******:****..***:**********::************: ** **
Mflv_2971|M.gilvum_PYR-GCK YETACDPRLNTQQSLELAFLVAEMLRD
Mvan_3540|M.vanbaalenii_PYR-1 YETACDPRLNTQQSLELAFLVAEMLRG
MSMEG_4244|M.smegmatis_MC2_155 YETACDPRLNTQQSLELAFLVAEMLRD
TH_1102|M.thermoresistible__bu YETACDPRLNTQQSLELAFLVAEMLRD
MMAR_1854|M.marinum_M YETACDPRLNTQQSLELAFLVAEMLRN
MUL_3533|M.ulcerans_Agy99 YETACDPRLNTQQSLELAFLVAEMLRD
Mb2200c|M.bovis_AF2122/97 YETACDPRLNTQQSLELAFLVAEMLRD
Rv2178c|M.tuberculosis_H37Rv YETACDPRLNTQQSLELAFLVAEMLRD
MAV_2317|M.avium_104 YETACDPRLNTQQSLELAFLVAEMLRD
MLBr_00896|M.leprae_Br4923 YETACDPRLNTQQSLELAFLVAEMLRD
MAB_1987|M.abscessus_ATCC_1997 YETACDPRLNTQQSLELAFLVAEMLRD
**************************.