For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NWTVDIPIDQLPSLPPLPTDLRTRLDAALAKPAAQQPTWPADQALAMRTVLESVPPVTVPSEIVRLQEQL AQVAKGEAFLLQGGDCAETFMDNTEPHIRGNVRALLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDA LGLRSYRGDMINGFAPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDWNREFVRTSPAG ARYEALATEIDRGLRFMSACGVADRNLQTAEIYASHEALVLDYERAMLRLSDGDDGEPQLFDLSAHTVWI GERTRQIDGAHIAFAQVIANPVGVKLGPNMTPELAVEYVERLDPHNKPGRLTLVSRMGNHKVRDLLPPIV EKVQATGHQVIWQCDPMHGNTHESSTGFKTRHFDRIVDEVQGFFEVHRALGTHPGGIHVEITGENVTECL GGAQDISETDLAGRYETACDPRLNTQQSLELAFLVAEMLRD*
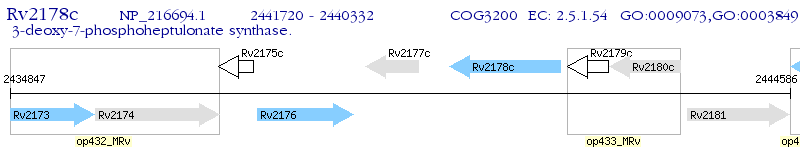
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2178c | aroG | - | 100% (462) | Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG (DAHP synthetase, phenylalanine-repressible) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2200c | aroG | 0.0 | 99.78% (462) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. gilvum PYR-GCK | Mflv_2971 | - | 0.0 | 86.32% (468) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. leprae Br4923 | MLBr_00896 | aroG | 0.0 | 89.68% (465) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAHP |
| M. abscessus ATCC 19977 | MAB_1987 | - | 0.0 | 85.93% (462) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. marinum M | MMAR_3222 | aroG | 0.0 | 91.99% (462) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG |
| M. avium 104 | MAV_2317 | - | 0.0 | 93.51% (462) | 3-deoxy-7-phosphoheptulonate synthase |
| M. smegmatis MC2 155 | MSMEG_4244 | - | 0.0 | 87.07% (464) | 3-deoxy-7-phosphoheptulonate synthase |
| M. thermoresistible (build 8) | TH_1102 | aroG | 0.0 | 87.88% (462) | Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate |
| M. ulcerans Agy99 | MUL_3533 | aroG_1 | 0.0 | 91.77% (462) | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_3540 | - | 0.0 | 86.30% (467) | phospho-2-dehydro-3-deoxyheptonate aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2971|M.gilvum_PYR-GCK MNWTVDVPIDQLPALPPLPADLRQRLDAALAKPALQQPSWDAGQAAAMRK
Mvan_3540|M.vanbaalenii_PYR-1 MNWTVDVPIDQLPALPPLPEDLRQRLDAALARPALQQPSWDAGQAAAMRK
MSMEG_4244|M.smegmatis_MC2_155 MNWTVDIPIDQLPPLPPLSDELRQRLDSALAKPAVQQPSWDPDAAKAMRT
TH_1102|M.thermoresistible__bu VNWTVDVPIDQLPALPPLPADLRQRLDAALAKPALQQPSWDPDLAKAMRT
Rv2178c|M.tuberculosis_H37Rv MNWTVDIPIDQLPSLPPLPTDLRTRLDAALAKPAAQQPTWPADQALAMRT
Mb2200c|M.bovis_AF2122/97 MNWTVDIPIDQLPSLPPLPTDLRTRLDAALAKPAAQQPTWPADQALAMRT
MAV_2317|M.avium_104 MNWTVDIPIDQLPPLPPLPGDLRARLDAALAKPAAQQPSWPTDQATAMRT
MMAR_3222|M.marinum_M MNWTVDIPIDQLPSLPPLSDELRARLDAALAKPAAQQPSWPVDQATAMRK
MUL_3533|M.ulcerans_Agy99 MNWTVDIPIDQLPSLPPLSDELRARLDAALAKPAAQQPSWPVDQATAMRK
MLBr_00896|M.leprae_Br4923 MNWTVDIPIDQLP---PLPADLRARLDAALAKPAAQQPSWPTDQAAAMRT
MAB_1987|M.abscessus_ATCC_1997 MNWTVDVPIDQLPELPPLPADLRERLDAALAKPAAQQPSWPANQAAAMRT
:*****:****** **. :** ***:***:** ***:* . * ***.
Mflv_2971|M.gilvum_PYR-GCK VLESVPPVTVPSEIERLKGQLADVALGKAFLLQGGDCAETFVDNTEPHIR
Mvan_3540|M.vanbaalenii_PYR-1 VLESVPPVTVPSEIERLKSQLADVARGQAFLLQGGDCAETFVDNTEPHIR
MSMEG_4244|M.smegmatis_MC2_155 VLESVPPVTVPSEIEKLKGLLADVAQGKAFLLQGGDCAETFVDNTEPHIR
TH_1102|M.thermoresistible__bu VLESVPPITVPSEVERLKSLLGEVARGKAFLLQGGDCAETFVDNTEPHIK
Rv2178c|M.tuberculosis_H37Rv VLESVPPVTVPSEIVRLQEQLAQVAKGEAFLLQGGDCAETFMDNTEPHIR
Mb2200c|M.bovis_AF2122/97 VLESVPPVTVPSEIVRLQEQLAQVAKGEAFLLQGGDCAETFMDNTEPHIR
MAV_2317|M.avium_104 VLESVPPVTVPSEIVRLQEQLAQVARGEAFLLQGGDCAETFTENTEPHIR
MMAR_3222|M.marinum_M VLESVPPVTVPSEIIRLQEQLAQVARGEAFLLQGGDCAETFTDNTEPHIR
MUL_3533|M.ulcerans_Agy99 VLESVPPVTVPSEIIRLQEQLAQVAGGEAFLLQGGDCAETFTDNTEPHIR
MLBr_00896|M.leprae_Br4923 VLESVPPVTVPSEIIRLQEQLALVANGKAFLLQGGDCAETFVDNTEPHIR
MAB_1987|M.abscessus_ATCC_1997 VLESVPPITVPAEIQRLQRQLAQVARGEAFLLQGGDCAETFADNTEPHIR
*******:***:*: :*: *. ** *:************* :******:
Mflv_2971|M.gilvum_PYR-GCK ANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDVDALGLKSYRGD
Mvan_3540|M.vanbaalenii_PYR-1 ANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDTDALGLKSYRGD
MSMEG_4244|M.smegmatis_MC2_155 ANIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSSDVDALGLKSYRGD
TH_1102|M.thermoresistible__bu ANIRTLLQMAVVLTYGASTPVVKMARIAGQYAKPRSSDTDALGLKSYRGD
Rv2178c|M.tuberculosis_H37Rv GNVRALLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDALGLRSYRGD
Mb2200c|M.bovis_AF2122/97 GNVRALLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDALGLRSYRGD
MAV_2317|M.avium_104 GNIRTLLQMAVVLTYGASMPVVKVARIAGQYAKPRSADIDALGLKSYRGD
MMAR_3222|M.marinum_M GNVRTLLQMAVVLTYGSSMPVVKVARIAGQYAKPRSADTDALGLKSYRGD
MUL_3533|M.ulcerans_Agy99 GNVRTLLQMAVVLTYGSSMPVVKVARIAGQYAKPRSADTDALGLKSYRGD
MLBr_00896|M.leprae_Br4923 SNVRTLLQMAVVLTYGASLPVVKVARIAGQYAKPRSADVDALGLKSYRGD
MAB_1987|M.abscessus_ATCC_1997 ANIRALLQMAVVLTYGASMPVVKLARIAGQYAKPRSSDTDALGLKSYRGD
.*:*:***********:* ****:************:* *****:*****
Mflv_2971|M.gilvum_PYR-GCK MVNGFAPDAAVRDHDPSRLVRAYANASAAMNLVRALTSSGMASLHQVHDW
Mvan_3540|M.vanbaalenii_PYR-1 MVNGFAPDAAVREHDPSRLVRAYANASAAMNLVRALTSSGMASLHQVHDW
MSMEG_4244|M.smegmatis_MC2_155 MINGFAPDAAAREHDPSRLVRAYANASAAMNLMRALTSSGLASLHLVHEW
TH_1102|M.thermoresistible__bu MINGFAPDASAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
Rv2178c|M.tuberculosis_H37Rv MINGFAPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
Mb2200c|M.bovis_AF2122/97 MINGFAPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
MAV_2317|M.avium_104 MINGFAPNAAVREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
MMAR_3222|M.marinum_M MINGFPPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
MUL_3533|M.ulcerans_Agy99 MINGFPPDAAAREHDPSRLVRAYANASAAMNLVRALTSSGLASLHLVHDW
MLBr_00896|M.leprae_Br4923 MINGFAPDAAAREHDPSRLVRAYANSSAAMNLVRALTSSGLASLHLVHDW
MAB_1987|M.abscessus_ATCC_1997 MVNGFAPDATLREHDPSRLVRAYANASAAMNLVRALTGSGMASLALVHDW
*:***.*:*: *:************:******:****.**:*** **:*
Mflv_2971|M.gilvum_PYR-GCK NREFVRTSPAGARYEALAGEIDRGLRFMSACRVDDRNLDTAEIYASHEAL
Mvan_3540|M.vanbaalenii_PYR-1 NREFVRTSPAGARYEALAGEIDRGLRFMSACRVDDRNLDTAEIYASHEAL
MSMEG_4244|M.smegmatis_MC2_155 NREFVRTSPAGARYEALAGEIDRGLNFMSACGVADRNLQTAEIFASHEAL
TH_1102|M.thermoresistible__bu NREFVRTSPAGARYEALAGEIDRGLRFMSACGVADRNLQTAEIYASHEAL
Rv2178c|M.tuberculosis_H37Rv NREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTAEIYASHEAL
Mb2200c|M.bovis_AF2122/97 NREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTAEIYASHEAL
MAV_2317|M.avium_104 NREFVRTSPAGARYEALATEIDRGLRFMSACGVADRNLQTAEIYASHEAL
MMAR_3222|M.marinum_M NREFVRTSPAGARYEALANEIDRGLRFMSACGVADRNLQTAEIYASHEAL
MUL_3533|M.ulcerans_Agy99 NREFVRTSPAGARYEALANEIDRGLRFMSACGVADRNLQTAEIYASHEAL
MLBr_00896|M.leprae_Br4923 NREFVRTSPAGARYEALAGEIGRGLAFMSACGVADRNLQTAEIYASHEAL
MAB_1987|M.abscessus_ATCC_1997 NREFVRTSPAGARYEALAGEIDRGLKFMSACGVVDSNLDTAEIYASHEAL
****************** **.*** ***** * * **:****:******
Mflv_2971|M.gilvum_PYR-GCK VLDYERAMLRMETGDLADPALSSEPKLYDLSAHYVWIGERTRQLDGAHVA
Mvan_3540|M.vanbaalenii_PYR-1 VLDYERAMLRMNIADSADPASDGPPKLYDLSAHYVWIGERTRQLDGAHVA
MSMEG_4244|M.smegmatis_MC2_155 VLDYERAMLRLSN----PAETDGAAKLYDQSAHYLWIGERTRQLDGAHVA
TH_1102|M.thermoresistible__bu VLDYERAMLRLS-------EETGEPKLYDLSAHYLWIGERTRQLDGAHIA
Rv2178c|M.tuberculosis_H37Rv VLDYERAMLRLS------DGDDGEPQLFDLSAHTVWIGERTRQIDGAHIA
Mb2200c|M.bovis_AF2122/97 VLDYERAMLRLS------DGEDGEPQLFDLSAHTVWIGERTRQIDGAHIA
MAV_2317|M.avium_104 VLDYERAMLRLS------DSDAGEPQLYDLSAHTVWIGERTRQLDGAHIA
MMAR_3222|M.marinum_M VLDYERAMLRLG------EGENGEPQLYDLSAHTVWIGERTRQLDGAHVA
MUL_3533|M.ulcerans_Agy99 VLDYERVMLRLG------EGENGEPQLYDLSAHTVWIGERTRQLDGAHVA
MLBr_00896|M.leprae_Br4923 VLDYERAMLRLAG---APEGPDDGLQLYDLSAHTVWIGERTRQLDGAHVA
MAB_1987|M.abscessus_ATCC_1997 VLDYERAMLRLG------EDENGEPALYDLSAHYLWIGDRTRQLDHAHVA
******.***: . *:* *** :***:****:* **:*
Mflv_2971|M.gilvum_PYR-GCK FAEVIANPIGIKIGPTTSPELAVEYVERLDPNNEPGRLTLVSRMGNHKVR
Mvan_3540|M.vanbaalenii_PYR-1 FAEVIANPVGIKIGPTTSPELAVEYVERLDPNNEPGRLTLVSRMGNHKVR
MSMEG_4244|M.smegmatis_MC2_155 FAEVIANPIGVKLGPTTTPELAVEYVERLDPNNEPGRLTLVTRMGNNKVR
TH_1102|M.thermoresistible__bu FAEVIANPIGVKIGPTTTPEVAAEYVERLDPNNEPGRLTLVSRLGNHKVR
Rv2178c|M.tuberculosis_H37Rv FAQVIANPVGVKLGPNMTPELAVEYVERLDPHNKPGRLTLVSRMGNHKVR
Mb2200c|M.bovis_AF2122/97 FAQVIANPVGVKLGPNMTPELAVEYVERLDPHNKPGRLTLVSRMGNHKVR
MAV_2317|M.avium_104 FAEVIANPIGVKIGPTITPELAVEYVERLDPHNKPGRLTLVSRMGNSKVR
MMAR_3222|M.marinum_M FAEVIANPIGVKIGPTMTPELAVEYVERLDPHNKAGRLTLVSRLGNNKVR
MUL_3533|M.ulcerans_Agy99 FAEVIANPIGVKIGPTMTPELAVEYVERLDPHNKAGRLTLVSRLGNNKVR
MLBr_00896|M.leprae_Br4923 FAEVIANPIGVKMGATMTPELAVEYVERLDPHNKPGRLTLVSRLGNNKVR
MAB_1987|M.abscessus_ATCC_1997 FAEIIANPIGMKIGPTTTPDQAVEYVERLDPHNKPGRLTFVSRMGNSKVR
**::****:*:*:*.. :*: *.********:*:.****:*:*:** ***
Mflv_2971|M.gilvum_PYR-GCK DVLPPIIEKVQASGHRVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
Mvan_3540|M.vanbaalenii_PYR-1 DVLPPIIEKVQASGHQVVWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
MSMEG_4244|M.smegmatis_MC2_155 DLLPPIIEKVQATGHQVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
TH_1102|M.thermoresistible__bu DLLPPIIEKVQATGHTVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
Rv2178c|M.tuberculosis_H37Rv DLLPPIVEKVQATGHQVIWQCDPMHGNTHESSTGFKTRHFDRIVDEVQGF
Mb2200c|M.bovis_AF2122/97 DLLPPIVEKVQATGHQVIWQCDPMHGNTHESSTGFKTRHFDRIVDEVQGF
MAV_2317|M.avium_104 DLLPPIVEKVQATGHQVIWQCDPMHGNTHESSNGYKTRHFDRIVDEVQGF
MMAR_3222|M.marinum_M DLLPPIVEKVQAAGHQVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
MUL_3533|M.ulcerans_Agy99 DLLPPIVEKVQAAGHQVIWQCDPMHGNTHESSTGYKTRHFDRIVDEVQGF
MLBr_00896|M.leprae_Br4923 DVLPPIVEKVKATGHQVIWQCDPMHGNTHESSTGYKTRHFDRVVDEVQGF
MAB_1987|M.abscessus_ATCC_1997 DLLPPIIEKVRATGHQVVWQCDPMHGNTHESPSGYKTRHFDRIVDEVQGY
*:****:***:*:** *:*************..*:*******:******:
Mflv_2971|M.gilvum_PYR-GCK FEVHRALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGRYETACDPRL
Mvan_3540|M.vanbaalenii_PYR-1 FEVHHALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGRYETACDPRL
MSMEG_4244|M.smegmatis_MC2_155 FEVHHALGTHPGGIHVEITGENVTECLGGAQDISDSDLAGRYETACDPRL
TH_1102|M.thermoresistible__bu FEVHRALGTHPGGIHVEITGENVTECLGGAQDISDTDLAGRYETACDPRL
Rv2178c|M.tuberculosis_H37Rv FEVHRALGTHPGGIHVEITGENVTECLGGAQDISETDLAGRYETACDPRL
Mb2200c|M.bovis_AF2122/97 FEVHRALGTHPGGIHVEITGENVTECLGGAQDISETDLAGRYETACDPRL
MAV_2317|M.avium_104 FEVHRALGTHPGGIHVEITGDNVTECLGGAQDISDMDLVGRYETACDPRL
MMAR_3222|M.marinum_M FEVHRGLGTHPGGIHVEITGENVTECLGGAQDISDSDLSGRYETACDPRL
MUL_3533|M.ulcerans_Agy99 FEVHRGLGTHPGGIHVEITGENVTECLGGAQDISDSDLSGRYETACDPRL
MLBr_00896|M.leprae_Br4923 FEVHRALGTYPGGIHVEITGEDVTECLGGAQDISDTDLAGRYETACDPRL
MAB_1987|M.abscessus_ATCC_1997 FEVHNALGTHPGGIHVEITGENVTECLGGAQDISDDDLAGRYETACDPRL
****..***:**********::************: ** ***********
Mflv_2971|M.gilvum_PYR-GCK NTQQSLELAFLVAEMLRD
Mvan_3540|M.vanbaalenii_PYR-1 NTQQSLELAFLVAEMLRG
MSMEG_4244|M.smegmatis_MC2_155 NTQQSLELAFLVAEMLRD
TH_1102|M.thermoresistible__bu NTQQSLELAFLVAEMLRD
Rv2178c|M.tuberculosis_H37Rv NTQQSLELAFLVAEMLRD
Mb2200c|M.bovis_AF2122/97 NTQQSLELAFLVAEMLRD
MAV_2317|M.avium_104 NTQQSLELAFLVAEMLRD
MMAR_3222|M.marinum_M NTQQSLELAFLVAEMLRD
MUL_3533|M.ulcerans_Agy99 NTQQSLELAFLVAEMLRD
MLBr_00896|M.leprae_Br4923 NTQQSLELAFLVAEMLRD
MAB_1987|M.abscessus_ATCC_1997 NTQQSLELAFLVAEMLRD
*****************.