For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSLFRRMWVPIVMILVAIVGASVVMRLHGVFGSHRYVPDAGNSDAIVAFNPKYVRYEVFGPPGTVVTVNY LDAQAEPHEVANAAIPWSLTIVTTLTSVVAHLVAQGNSATLSCRITVNGVVRAERTADAHDAATSCLVKS A*
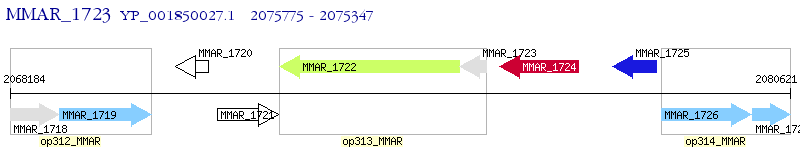
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1723 | mmpS1 | - | 100% (142) | membrane protein MmpS1 |
| M. marinum M | MMAR_0705 | mmpS1_1 | 3e-52 | 64.79% (142) | membrane protein MmpS1_1 |
| M. marinum M | MMAR_3049 | mmpS4_1 | 9e-37 | 49.65% (141) | membrane transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0410c | mmpS1 | 7e-51 | 62.68% (142) | membrane protein |
| M. gilvum PYR-GCK | Mflv_3434 | - | 3e-28 | 40.56% (143) | membrane family protein |
| M. tuberculosis H37Rv | Rv0403c | mmpS1 | 7e-51 | 62.68% (142) | membrane protein |
| M. leprae Br4923 | MLBr_02377 | - | 4e-35 | 47.86% (140) | hypothetical protein MLBr_02377 |
| M. abscessus ATCC 19977 | MAB_3149 | - | 3e-31 | 43.75% (144) | MmpS family protein |
| M. avium 104 | MAV_3864 | - | 1e-35 | 50.35% (143) | MmpS4 protein |
| M. smegmatis MC2 155 | MSMEG_0575 | - | 1e-32 | 46.76% (139) | MmpS1 protein |
| M. thermoresistible (build 8) | TH_2866 | - | 1e-31 | 43.80% (137) | PUTATIVE hypothetical protein Mvan_3834 |
| M. ulcerans Agy99 | MUL_1965 | mmpS1 | 4e-73 | 98.52% (135) | membrane protein MmpS1 |
| M. vanbaalenii PYR-1 | Mvan_1058 | - | 1e-31 | 42.25% (142) | hypothetical protein Mvan_1058 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1723|M.marinum_M -----------MVSLFRRMWVPIVMILVAIVGASVVMRLHGVFGSHRYVP
MUL_1965|M.ulcerans_Agy99 ------------------MWVPIVMVLVAIVGASVVMRLHGVFGSHRYVP
Mb0410c|M.bovis_AF2122/97 -----------MFGVAKRFWIPMVIVIVVAVAAVTVSRLHSVFGSHQHAP
Rv0403c|M.tuberculosis_H37Rv -----------MFGVAKRFWIPMVIVIVVAVAAVTVSRLHSVFGSHQHAP
MSMEG_0575|M.smegmatis_MC2_155 --------------MLKQAWIPLVLAVVLPVSGLVVWRLHEKFGSEDLNA
MLBr_02377|M.leprae_Br4923 MSKRQRGKGISIFKLLSRIWIPLVILVVLVVGGFVVYRVHSYFASEKRES
MAV_3864|M.avium_104 ------MKGFSIASLVKRGWMVLVVVVVVGVAGFCVYRLHGIFGSHNNTS
TH_2866|M.thermoresistible__bu -LWTSKAKDSAILTRLARAWIPLVMVVVVAIGGFAVWRIRGIFGSEQLPT
MAB_3149|M.abscessus_ATCC_1997 -----------MFRTIRKIWMPLLIAAVVAIGGFAVFRVRGIFGSESFGA
Mflv_3434|M.gilvum_PYR-GCK -----------MIAIGKKVWLPLLIVVVVVIAGATVSRIRTFFGSEGIIE
Mvan_1058|M.vanbaalenii_PYR-1 -----------MITVVKRVWLPVLVIAAVAVGAVTVANLRSVFGSDKAIV
*: ::: . :.. * .:: *.*.
MMAR_1723|M.marinum_M DAG-NSDAIVAFNPKYVRYEVFGPPGTVVTVNYLDAQAEPHEVANAAIPW
MUL_1965|M.ulcerans_Agy99 DAG-NSDAIVAFNPKYVRYEVFGPPGTVVTVNYLDAQAEPHEVANAAIPW
Mb0410c|M.bovis_AF2122/97 DTG-NLDPIIAFYPKHVLYEVFGPPGTVASINYLDADAQPHEVVNAAVPW
Rv0403c|M.tuberculosis_H37Rv DTG-NLDPIIAFYPKHVLYEVFGPPGTVASINYLDADAQPHEVVNAAVPW
MSMEG_0575|M.smegmatis_MC2_155 NAG-AGIEIVQFNPKILVYEVYGPPGSTAQISYFDTDANVHS-VNTTLPW
MLBr_02377|M.leprae_Br4923 YADSNLGSSKPFNPKQIVYEVFGPPGTVADISYFDANSDPQRIDGAQLPW
MAV_3864|M.avium_104 AAGGISDQTEPFSPKHITLEVFGNPGAVATINYVDINVQPRQVLNAALPW
TH_2866|M.thermoresistible__bu YAGSTSVDTDKANPKRVRYEIFGAPGTVADINYIDAEGDPHQVNGATLPW
MAB_3149|M.abscessus_ATCC_1997 SADNNAKDAVPYNPKTLVYEIFGEPGAMADVNYFNEDAQPTRVDNVQLPW
Mflv_3434|M.gilvum_PYR-GCK TPRNFADLPEPFDPKVVRYEIIGN-GSYADVNYLDLDAKPQRVDGAALPW
Mvan_1058|M.vanbaalenii_PYR-1 TPVD-ADTAENFNPKVVTYEIFGS-GSTATINYADLDGKPQRTGEVSLPW
. ** : *: * *: . :.* : : . . :**
MMAR_1723|M.marinum_M SLTIVTTLTSVVAHLVAQGNSATLSCRITVNGVVRAERTADAHDAATSCL
MUL_1965|M.ulcerans_Agy99 SLTIVTTLTSVVAHLVAQGNSATLSCRITVNGVVRAERTADAHDAATSCP
Mb0410c|M.bovis_AF2122/97 SFTIVTTLTAVVANVVARGDGASLGCRITVNEVIREERIVNAYHAHTSCL
Rv0403c|M.tuberculosis_H37Rv SFTIVTTLTAVVANVVARGDGASLGCRITVNEVIREERIVNAYHAHTSCL
MSMEG_0575|M.smegmatis_MC2_155 SVTLSTTLPTVSANLMALGSNDEIGCRVTVNGIVREEKSANGIDAQTYCL
MLBr_02377|M.leprae_Br4923 SLLMTTTLAAVMGNLVAQGNTDSIGCRIIVDGVVKAERVSNEVNAYTYCL
MAV_3864|M.avium_104 SLTMVTTQPGAFSNLVAQGNSDSLGCRITVDGDVKDERIVNQVNAYTFCL
TH_2866|M.thermoresistible__bu SIEIVTNSPAMAGNVLAQGDSDFIGCRIVSDGVVKDERTTQRVSAYVYCF
MAB_3149|M.abscessus_ATCC_1997 AFKIVSTLPSLSGNIVAQGNGDQIGCRITVNGEVKMERVSTEHNAYIYCL
Mflv_3434|M.gilvum_PYR-GCK SLTLETTEPSAAPNIVAQGDSSTITCRIFVDDELKDERTTDGLNAQTFCF
Mvan_1058|M.vanbaalenii_PYR-1 SLTLETTLPSVMPNIMAQGDGQSITCRVTVDDEVKDERTAEGLNAATYCL
:. : :. . :::* *. : **: : :: *: * *
MMAR_1723|M.marinum_M VKSA
MUL_1965|M.ulcerans_Agy99 VKSA
Mb0410c|M.bovis_AF2122/97 VKSA
Rv0403c|M.tuberculosis_H37Rv VKSA
MSMEG_0575|M.smegmatis_MC2_155 VKSA
MLBr_02377|M.leprae_Br4923 VKSA
MAV_3864|M.avium_104 VKSA
TH_2866|M.thermoresistible__bu TKSA
MAB_3149|M.abscessus_ATCC_1997 VKSA
Mflv_3434|M.gilvum_PYR-GCK VKSA
Mvan_1058|M.vanbaalenii_PYR-1 VKAA
.*:*