For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KGFSIASLVKRGWMVLVVVVVVGVAGFCVYRLHGIFGSHNNTSAAGGISDQTEPFSPKHITLEVFGNPGA VATINYVDINVQPRQVLNAALPWSLTMVTTQPGAFSNLVAQGNSDSLGCRITVDGDVKDERIVNQVNAYT FCLVKSA*
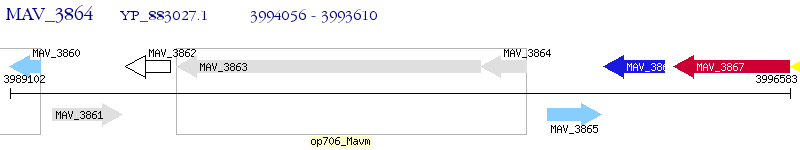
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3864 | - | - | 100% (148) | MmpS4 protein |
| M. avium 104 | MAV_3247 | - | 3e-36 | 51.47% (136) | MmpS4 protein |
| M. avium 104 | MAV_2511 | - | 1e-33 | 46.15% (143) | MmpS5 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1582 | mmpS6 | 2e-57 | 67.57% (148) | membrane protein |
| M. gilvum PYR-GCK | Mflv_3434 | - | 2e-33 | 46.15% (143) | membrane family protein |
| M. tuberculosis H37Rv | Rv0451c | mmpS4 | 4e-42 | 54.29% (140) | membrane protein |
| M. leprae Br4923 | MLBr_02377 | - | 5e-42 | 55.10% (147) | hypothetical protein MLBr_02377 |
| M. abscessus ATCC 19977 | MAB_3149 | - | 6e-36 | 46.04% (139) | MmpS family protein |
| M. marinum M | MMAR_1696 | - | 2e-53 | 65.00% (140) | hypothetical protein MMAR_1696 |
| M. smegmatis MC2 155 | MSMEG_3495 | - | 5e-41 | 53.06% (147) | MmpS5 protein |
| M. thermoresistible (build 8) | TH_2866 | - | 3e-37 | 48.30% (147) | PUTATIVE hypothetical protein Mvan_3834 |
| M. ulcerans Agy99 | MUL_1935 | - | 7e-50 | 64.66% (133) | hypothetical protein MUL_1935 |
| M. vanbaalenii PYR-1 | Mvan_3834 | - | 1e-35 | 47.48% (139) | hypothetical protein Mvan_3834 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1696|M.marinum_M ---------------MEALVRRGWMMIVAVAVVVIAGFAIYRLHGIFGSH
MUL_1935|M.ulcerans_Agy99 ------------------------MMIVAVAVVVIAGFAIYRLHGIFGSH
Mb1582|M.bovis_AF2122/97 ----------MQGISVTGLVKRGWMVLVAVAVVAVAGFSVYRLHGIFGSH
MAV_3864|M.avium_104 ----------MKGFSIASLVKRGWMVLVVVVVVGVAGFCVYRLHGIFGSH
TH_2866|M.thermoresistible__bu -----LWTSKAKDSAILTRLARAWIPLVMVVVVAIGGFAVWRIRGIFGSE
Mvan_3834|M.vanbaalenii_PYR-1 -------------------MTRAWIPLVLVLVLVVGGLAVWRIRNIFGSH
Rv0451c|M.tuberculosis_H37Rv ------------------MLMRTWIPLVILVVVIVGGFTVHRIRGFFGSE
MLBr_02377|M.leprae_Br4923 ----MSKRQRGKGISIFKLLSRIWIPLVILVVLVVGGFVVYRVHSYFASE
Mflv_3434|M.gilvum_PYR-GCK ---------------MIAIGKKVWLPLLIVVVVVIAGATVSRIRTFFGSE
MSMEG_3495|M.smegmatis_MC2_155 MMAVDTKDKQRKKFSISALARRFWIPLLIGAVVFVGAFTVMRVRTFFGSG
MAB_3149|M.abscessus_ATCC_1997 ---------------MFRTIRKIWMPLLIAAVVAIGGFAVFRVRGIFGSE
: :: *: :.. : *:: *.*
MMAR_1696|M.marinum_M NNASGANGISNEIVPFNPKQVVLDVFGAPGAVATINYLDVNAQPQQVVDA
MUL_1935|M.ulcerans_Agy99 NNASGANGISNEIVPFNPKQVVLDVFGAPGAVATINYLDVNAQPQQVVDA
Mb1582|M.bovis_AF2122/97 DTTSTAGGVANDIKPFNPKQVTLEVFGAPGTVATINYLDVDATPRQVLDT
MAV_3864|M.avium_104 NNTSAAGGISDQTEPFSPKHITLEVFGNPGAVATINYVDINVQPRQVLNA
TH_2866|M.thermoresistible__bu QLPTYAGSTSVDTDKANPKRVRYEIFGAPGTVADINYIDAEGDPHQVNGA
Mvan_3834|M.vanbaalenii_PYR-1 QLPTYAGSMSEDSNKSDPKIVRYEIFGDPGATADINYIDDEGDPNQIVGA
Rv0451c|M.tuberculosis_H37Rv NRPSYSDTNLENSKPFNPKHLTYEIFGPPGTVADISYFDVNSEPQRVDGA
MLBr_02377|M.leprae_Br4923 KRESYADSNLGSSKPFNPKQIVYEVFGPPGTVADISYFDANSDPQRIDGA
Mflv_3434|M.gilvum_PYR-GCK GIIETPRNFADLPEPFDPKVVRYEIIGN-GSYADVNYLDLDAKPQRVDGA
MSMEG_3495|M.smegmatis_MC2_155 DDPSLASAKVDDTKPFDPKVVVYEIYGEPGATADINYLDLDAQPQRVDGA
MAB_3149|M.abscessus_ATCC_1997 SFGASADNNAKDAVPYNPKTLVYEIFGEPGAMADVNYFNEDAQPTRVDNV
. .** : :: * *: * :.*.: : * :: ..
MMAR_1696|M.marinum_M PLPWSFTITTTEPAVLGNVVAQGNGGTIGCRITVNGEVKDERIVNTPDAY
MUL_1935|M.ulcerans_Agy99 PLPWSFTITTTEPAVLGNMVAQGNGGTIGCRITVNGEVKDERIVNTPDAY
Mb1582|M.bovis_AF2122/97 TLPWSYTITTTLPAVFANVVAQGDSNSIGCRITVNGVVKDERIVNEVRAY
MAV_3864|M.avium_104 ALPWSLTMVTTQPGAFSNLVAQGNSDSLGCRITVDGDVKDERIVNQVNAY
TH_2866|M.thermoresistible__bu TLPWSIEIVTNSPAMAGNVLAQGDSDFIGCRIVSDGVVKDERTTQRVSAY
Mvan_3834|M.vanbaalenii_PYR-1 TLPWSIEVETNSPAMVGNVVAQGDSDFIGCRISADGVVKDERTAHNVSAY
Rv0451c|M.tuberculosis_H37Rv VLPWSLHITTNDAAVMGNIVAQGNSDSIGCRITVDGKVRAERVSNEVNAY
MLBr_02377|M.leprae_Br4923 QLPWSLLMTTTLAAVMGNLVAQGNTDSIGCRIIVDGVVKAERVSNEVNAY
Mflv_3434|M.gilvum_PYR-GCK ALPWSLTLETTEPSAAPNIVAQGDSSTITCRIFVDDELKDERTTDGLNAQ
MSMEG_3495|M.smegmatis_MC2_155 TLPWSLTLSTTAPSVFPNIVAQGNGDTITCRITVDDELKDERTSNGVNAQ
MAB_3149|M.abscessus_ATCC_1997 QLPWAFKIVSTLPSLSGNIVAQGNGDQIGCRITVNGEVKMERVSTEHNAY
***: : :. .. *::***: . : *** :. :: ** *
MMAR_1696|M.marinum_M TFCLDKSG
MUL_1935|M.ulcerans_Agy99 TFCLDKSG
Mb1582|M.bovis_AF2122/97 TFCLDKSS
MAV_3864|M.avium_104 TFCLVKSA
TH_2866|M.thermoresistible__bu VYCFTKSA
Mvan_3834|M.vanbaalenii_PYR-1 VYCFTKSA
Rv0451c|M.tuberculosis_H37Rv TYCLVKSA
MLBr_02377|M.leprae_Br4923 TYCLVKSA
Mflv_3434|M.gilvum_PYR-GCK TFCFVKSA
MSMEG_3495|M.smegmatis_MC2_155 TFCLVKSA
MAB_3149|M.abscessus_ATCC_1997 IYCLVKSA
:*: **.