For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAEQAMYEQVNSYVVDPADVGSRIDPVLARSWLLVNGTHADRFQAASHSRADIVVLDIEDAVAPKDKGS ARDNVVHWLGDGNADWVRINGFGTPWWADDIAILAGSSVGGVMLAMVESVDHVTETARRLPNVPIVALVE TARGLERITEIASAKGTFRLAFGIGDFRRDTGFGEDATTLAYARSRFTIAAKAAGLPSAIDGPTIGSNPL KLIEASAVSVEFGMTGKICLSPEQCAVVNEGLSPSQDEIGWAKEFFAEFERDGGEIRNGSDLPRIARATK ILDLARAYGIEASDFEDEPVHSPAPTDTYHY
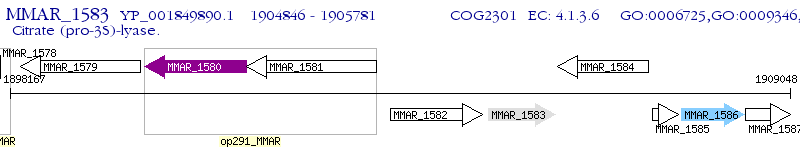
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1583 | citE_2 | - | 100% (311) | citrate lyase beta subunit, CitE_2 |
| M. marinum M | MMAR_3845 | citE | 2e-20 | 30.29% (241) | citrate (pro-3s)-lyase (beta subunit) CitE |
| M. marinum M | MMAR_4158 | citE_1 | 2e-18 | 27.72% (285) | citrate (pro-3s)-lyase (beta subunit) CitE_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3102c | - | 1e-155 | 87.58% (306) | hypothetical protein Mb3102c |
| M. gilvum PYR-GCK | Mflv_4009 | - | 1e-141 | 83.56% (292) | HpcH/HpaI aldolase |
| M. tuberculosis H37Rv | Rv3075c | - | 1e-155 | 87.58% (306) | hypothetical protein Rv3075c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4737 | - | 8e-58 | 42.01% (269) | putative citrate lyase/aldolase |
| M. avium 104 | MAV_3981 | - | 1e-150 | 84.97% (306) | HpcH/HpaI aldolase/citrate lyase family protein, putative |
| M. smegmatis MC2 155 | MSMEG_2667 | - | 1e-139 | 81.14% (297) | HpcH/HpaI aldolase/citrate lyase family protein, putative |
| M. thermoresistible (build 8) | TH_0719 | - | 5e-22 | 29.81% (265) | HpcH/HpaI aldolase/citrate lyase family protein |
| M. ulcerans Agy99 | MUL_3451 | citE_2 | 1e-179 | 99.36% (311) | citrate lyase beta subunit, CitE_2 |
| M. vanbaalenii PYR-1 | Mvan_2378 | - | 1e-140 | 83.22% (292) | HpcH/HpaI aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4009|M.gilvum_PYR-GCK ------MYDQSFGEAAFGE-QGQRIDPVLARSWLLVNGAHYERFQAAAHS
Mvan_2378|M.vanbaalenii_PYR-1 ------MYEQNFSESAFGE-QGQRIDPVLARSWLLVNGSHYDRFQAAAHS
MMAR_1583|M.marinum_M MAAEQAMYEQVNSYVVDPADVGSRIDPVLARSWLLVNGTHADRFQAASHS
MUL_3451|M.ulcerans_Agy99 MAAEQAMYEQVNSYVVDPADVGSRIDPVLARSWLLVNGTHADRFQAASHS
Mb3102c|M.bovis_AF2122/97 ---MTSMYEQVDTNTADPV-AGSRIDPVLARSWLLVNGAHGDRFESAAHS
Rv3075c|M.tuberculosis_H37Rv ---MTSMYEQVDTNTADPV-AGSRIDPVLARSWLLVNGAHGDRFESAAHS
MAV_3981|M.avium_104 ------MYEQVNSSAAEPDDIGSRIDPVLARSWLLVNGTHPDRFQSAVDS
MSMEG_2667|M.smegmatis_MC2_155 ------MYDQPIHDPDPSASPGFRIDPVLARSWLLVNGAQYERFAPASRS
MAB_4737|M.abscessus_ATCC_1997 ------------------------MNSKVSRSWLLMNPSREASLEAAIAS
TH_0719|M.thermoresistible__bu -----------------------MDLSAVGPAWLFCPADRPERFEKAAAA
. :. :**: : : * :
Mflv_4009|M.gilvum_PYR-GCK R-ADIVVLDIEDAVAPKDKAAARANVVRWLGEGHADWVRVNGFGTEWWAG
Mvan_2378|M.vanbaalenii_PYR-1 R-ADIVVLDIEDAVAPKDKAAARANVTRWFGEGHDDWVRINGFGTEWWAG
MMAR_1583|M.marinum_M R-ADIVVLDIEDAVAPKDKGSARDNVVHWLGDGNADWVRINGFGTPWWAD
MUL_3451|M.ulcerans_Agy99 R-ADIVVVDIEDAVAPKDKGSARDNVVHWLGDGNADWVRINGFGTPWWAD
Mb3102c|M.bovis_AF2122/97 R-ADIVVLDIEDAVAPKDKHAARDNAVRWFGDGNADWVRINGFGTPWWAD
Rv3075c|M.tuberculosis_H37Rv R-ADIVVLDIEDAVAPKDKHAARDNAVRWFGDGNADWVRINGFGTPWWAD
MAV_3981|M.avium_104 R-ADIVVFDIEDAVAPKDKNAARDNVVSWLRAGNVDWVRINGFGTPWWAD
MSMEG_2667|M.smegmatis_MC2_155 R-ADIVILDIEDAVAPKDKAAARDNVVRWLSEGNTDWVRVNGFGTPWWAD
MAB_4737|M.abscessus_ATCC_1997 TNADEVILDLEDAVAPNEKEPARGRVLEYLNSGGNAWVRVNDASSDFWSA
TH_0719|M.thermoresistible__bu A--DVVILDLEDGVAAKDRPAAREALLSTPLDPARTVVRVNPAGTADHRL
* *:.*:**.**.::: .** **:* .:
Mflv_4009|M.gilvum_PYR-GCK DLEMLAGTS-VGGVMLAMVESVDHVTETAKRLP-GVPIVALVETARGLER
Mvan_2378|M.vanbaalenii_PYR-1 DLEMLAGTS-VGGVMLAMVESVDHVTETVKRLP-GVPIVALVETARGLER
MMAR_1583|M.marinum_M DIAILAGSS-VGGVMLAMVESVDHVTETARRLP-NVPIVALVETARGLER
MUL_3451|M.ulcerans_Agy99 DIAILAGSS-VGGVMLAMVESVDHVTETARRLP-NVPIVALVETARGLER
Mb3102c|M.bovis_AF2122/97 DLAMLADSP-VGGVMLAMVESVDHVTETAKRLP-NVPIVALVETARGLER
Rv3075c|M.tuberculosis_H37Rv DLAMLADSP-VGGVMLAMVESVDHVTETAKRLP-NVPIVALVETARGLER
MAV_3981|M.avium_104 DLTALAGTP-VGGVMLAMVESVDHVTETAKRLP-DVPIVALVETARGLER
MSMEG_2667|M.smegmatis_MC2_155 DLDTLKGTS-VGGIMLAMVESVDHVTETANRLP-DVPIVALVETARGLER
MAB_4737|M.abscessus_ATCC_1997 DCRALADARGLKGVVLAKTEGGNHMWDTSNRLGGETPVIALIETARGLSR
TH_0719|M.thermoresistible__bu DLDALTRTP-YRTVMLAKTDAAEQVRALA-----PLQVVVLIETPLGALN
* * : ::** .:. ::: ::.*:**. * .
Mflv_4009|M.gilvum_PYR-GCK ITEIAAAKGTFRLAFGIGDFRRDTG-------FGGDALTLAYTRSRFTIA
Mvan_2378|M.vanbaalenii_PYR-1 INEIASAKGTFRLAFGIGDFRRDTG-------FGDNPMTLAYARSRFTIA
MMAR_1583|M.marinum_M ITEIASAKGTFRLAFGIGDFRRDTG-------FGEDATTLAYARSRFTIA
MUL_3451|M.ulcerans_Agy99 ITEIASAKGTFRLAFGIGDFRRDTG-------FGKDATTLAYARSRFTIA
Mb3102c|M.bovis_AF2122/97 INEIAAAKGTFRLAFGIGDFRRDTG-------FGEDPATLAYARSRFTIA
Rv3075c|M.tuberculosis_H37Rv INEIAAAKGTFRLAFGIGDFRRDTG-------FGEDPATLAYARSRFTIA
MAV_3981|M.avium_104 ITEIAATKGTFRLAFGIGDFRRDTG-------FGEDPATLAYTRSRFTIA
MSMEG_2667|M.smegmatis_MC2_155 ITEIAATKGTFRLAFGIGDFRRDTG-------FGENPATLAYARSRFTIA
MAB_4737|M.abscessus_ATCC_1997 VHDIAAARPCFRIAFGVNDYTLDIG-------VDQDPLALAYSRSQLVVA
TH_0719|M.thermoresistible__bu VVESARADNAVAVMWGAEDLFAVTGGTDNRNPDGSYRDVARHVRSQSLLA
: : * : . : :* * * . . : **: :*
Mflv_4009|M.gilvum_PYR-GCK AKAAHLPSAIDGPTVGSSALKLSEATAVSAEFGMTGKICLTPEQCLTVNE
Mvan_2378|M.vanbaalenii_PYR-1 AKAAHLPSAIDGPTVGSSALRLSEATAVSAEFGMTGKICLTPEQCPTVNE
MMAR_1583|M.marinum_M AKAAGLPSAIDGPTIGSNPLKLIEASAVSVEFGMTGKICLSPEQCAVVNE
MUL_3451|M.ulcerans_Agy99 AKAAGLPSAIDGPTIGSNPLKLIEASAVSVEFGMTGKICLSPEQCAVVNE
Mb3102c|M.bovis_AF2122/97 ARAAGLPSAIDGPTIGSNALKLIEATAVSAEFGMTGKICLSPDQCPVVNE
Rv3075c|M.tuberculosis_H37Rv ARAAGLPSAIDGPTIGSNALKLIEATAVSAEFGMTGKICLSPDQCPVVNE
MAV_3981|M.avium_104 SKAADLPSAIDGPTIGSNPLKLIEATAVSTQFGMTGKICLTPDQCSVVNE
MSMEG_2667|M.smegmatis_MC2_155 AKAAHLPSAIDGPTVGSSPLKLAEATAVSAEFGMTGKICLTPDQCPTVNE
MAB_4737|M.abscessus_ATCC_1997 SRAAHIPGPIDGPTR--DPAELPEAVSLTRRMGMTGKLALRSSDADTINT
TH_0719|M.thermoresistible__bu AKAHGRLALDSVYLDIKDLDGLRAEVDDAVAVGFDAKVAIHPTQVAVIRE
::* . . . * : .*: .*:.: . : .:.
Mflv_4009|M.gilvum_PYR-GCK GLSPSAEEIAWAKEFFVEFERDGGEIRNG---SDLPRIARANKILELARA
Mvan_2378|M.vanbaalenii_PYR-1 GLSPSAEEITWAKEFFVEFERDGGEIRNG---SDLPRIARATKILDLARA
MMAR_1583|M.marinum_M GLSPSQDEIGWAKEFFAEFERDGGEIRNG---SDLPRIARATKILDLARA
MUL_3451|M.ulcerans_Agy99 GLSPSQDEIGWAKEFFAEFERDGGEIRNG---SDLPRIARATKILDLARA
Mb3102c|M.bovis_AF2122/97 GLSPSQDEIVWAKEFFAEFARDGGEIRNG---SDLPRIARATKILDLARA
Rv3075c|M.tuberculosis_H37Rv GLSPSQDEIVWAKEFFAEFARDGGEIRNG---SDLPRIARATKILDLARA
MAV_3981|M.avium_104 GLSPSQDEIAWAKEFFAEFERDGGEIRNG---SDLPRIARATKILELARA
MSMEG_2667|M.smegmatis_MC2_155 GLSPSQDEISWAKEFFAEFQRDGGEIRNG---SDLPRIARANKILDLARA
MAB_4737|M.abscessus_ATCC_1997 GLSPSDEDVSWAQDFVDRFNSQGGKPKDG---SDLPRLNRARIILERARM
TH_0719|M.thermoresistible__bu GYTPTPDQVRWARHVLAAARDERGVFQFEGIMVDAPVLRRAERIVQLAPD
* :*: ::: **:... : * : * * : ** *:: *
Mflv_4009|M.gilvum_PYR-GCK YGIEPSEFDDVDEPTHIPAPSDTYHY
Mvan_2378|M.vanbaalenii_PYR-1 YGIEPSDFDDVDEPAHIPAPSDTYHY
MMAR_1583|M.marinum_M YGIEASDFEDEP--VHSPAPTDTYHY
MUL_3451|M.ulcerans_Agy99 YGIEASDFEDEP--VHSPAPTDTYHY
Mb3102c|M.bovis_AF2122/97 YGIEVSDFEDEP--VHMPAPTDTYHY
Rv3075c|M.tuberculosis_H37Rv YGIEVSDFEDEP--VHMPAPTDTYHY
MAV_3981|M.avium_104 YGIEALDIDEEER-DHSPAPSDTYHY
MSMEG_2667|M.smegmatis_MC2_155 YGIHEPEYSNDDP-DHVPAPSDTYHY
MAB_4737|M.abscessus_ATCC_1997 LGLITP--------------------
TH_0719|M.thermoresistible__bu PGR-----------------------
*