For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTYHRLMNTSGSVGKVGDLAGDACCDVSTAPAHTGHSGDAAWQQGVRWARRLAWISLAVLVTEGAVGVWE GVAVGSVALTGWALGGASEGLASAMVLWRFTGSRTLSETAERRAQRGVAVSFWLVAPLVAAESIRHLTDK HEVGTSVIGIGLTAFALVVMPILGRANHKLGVLLQSGATEGEGTQNYLCAGQAAGVLLGLAITAAWPGGW WIDPGIGLVIAGIAVWQGVRAWRGQDCGC
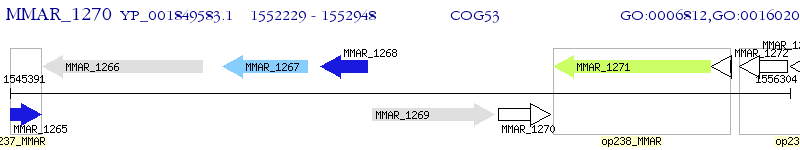
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1270 | - | - | 100% (239) | hypothetical protein MMAR_1270 |
| M. marinum M | MMAR_2136 | - | 5e-25 | 36.13% (191) | metal transport protein |
| M. marinum M | MMAR_3313 | - | 3e-10 | 32.28% (189) | hypothetical protein MMAR_3313 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3299c | - | 6e-91 | 73.36% (214) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3901 | - | 9e-25 | 37.50% (192) | Co/Zn/Cd cation transporters-like protein |
| M. tuberculosis H37Rv | Rv3271c | - | 6e-91 | 73.36% (214) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2603c | - | 6e-25 | 37.50% (184) | integral membrane protein |
| M. avium 104 | MAV_3555 | - | 2e-11 | 33.69% (187) | cation efflux family protein |
| M. smegmatis MC2 155 | MSMEG_5604 | - | 3e-26 | 38.86% (193) | integral membrane protein |
| M. thermoresistible (build 8) | TH_1241 | - | 1e-26 | 36.22% (196) | integral membrane protein |
| M. ulcerans Agy99 | MUL_2617 | - | 1e-134 | 98.71% (233) | hypothetical protein MUL_2617 |
| M. vanbaalenii PYR-1 | Mvan_3635 | - | 2e-69 | 52.70% (241) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1270|M.marinum_M MTYHRLMNTSGSVGKVGDLAGDACCDVSTAPAHTGHSGDAAWQQGVRWAR
MUL_2617|M.ulcerans_Agy99 ------MNTSSSVGKVGDLAGDACCDVSTTAAHTGHSGDAAWQQGVRWAR
Mb3299c|M.bovis_AF2122/97 ------METT-----------TEHRDESTLDSPVSVAREAEWQRNVRWAR
Rv3271c|M.tuberculosis_H37Rv ------METT-----------TEHRDESTLDSPVSVAREAEWQRNVRWAR
Mvan_3635|M.vanbaalenii_PYR-1 MTEIRAVTTPN-----LGSTSADCAADCCAAITTPVVRDAGWHRAARQAR
MSMEG_5604|M.smegmatis_MC2_155 -----------------MRRPRWRRVRMRMMADTSKLTPTRRAVLARRIR
TH_1241|M.thermoresistible__bu -------------------------VTPSGAIGWRPLSESRRQQLARRIR
MAB_2603c|M.abscessus_ATCC_199 ----------------------------------MVLAQQRRDVLNRRIR
Mflv_3901|M.gilvum_PYR-GCK ------MTDPAAEELKPLIASSAGCTDAGCTATAATVSPARRAVLTRRVR
MAV_3555|M.avium_104 --------------------MHVINGVRDPAASFPLDTATDDAGERRQAN
* .
MMAR_1270|M.marinum_M RLAWISLAVLVTEGAVGVWEGVAVGSVALTGWALGGASEGLASAMVLWRF
MUL_2617|M.ulcerans_Agy99 RLAWISLAVLVTEGAVGVWEGVAVGSVALTGWALGGASEGLASAMVLWRF
Mb3299c|M.bovis_AF2122/97 WLAWVSLAVLLTEGAVGLWQGIAVGSVALTGWALGGGSEGLASAMVLWRF
Rv3271c|M.tuberculosis_H37Rv WLAWVSLAVLLTEGAVGLWQGIAVGSVALTGWALGGGSEGLASAMVLWRF
Mvan_3635|M.vanbaalenii_PYR-1 LLAWVSLVWMGAEGAIGLWQGFTVGSIALIGWASGSAVEGLASIIVVWRF
MSMEG_5604|M.smegmatis_MC2_155 MFVAATITYNVIEAVVALTEGARVSSTALIGFGLDSVIEVSSAAAVAWQF
TH_1241|M.thermoresistible__bu VLVAATISYNIIEAVVAIGEGARVSSTALIGFGLDSAIEVSSAAAVAWQF
MAB_2603c|M.abscessus_ATCC_199 FFVVATIAYNVIEAIVALTEGSRVSSTALIGFGLDSVIEVSSAAAVAWQF
Mflv_3901|M.gilvum_PYR-GCK LFVLATITYNLIEAVVAISAGTIASSAALIGFGLDSLIEVASATAVAWQF
MAV_3555|M.avium_104 RAVAVSAAGLALTGLVELVIAVVSGSVALLGDALHNLSDVSTSALVFVGF
. : . : : . .* ** * . . : :: * *
MMAR_1270|M.marinum_M TGSRTL--------SETAERRAQRGVAVSFWLVAPLVAAESIRHLTDKHE
MUL_2617|M.ulcerans_Agy99 TGSRTL--------SETAERRAQRGVAVSFWLVAPLVAAESIRHLTDKHE
Mb3299c|M.bovis_AF2122/97 TGDRTW--------SATAEHRAQRGVAVSFWLTAPYLVAESIRHLAGEHR
Rv3271c|M.tuberculosis_H37Rv TGDRTW--------SATAEHRAQRGVAVSFWLTAPYLVAESIRHLAGEHR
Mvan_3635|M.vanbaalenii_PYR-1 TGAWTL--------SETAERRSQQAVAVSFWLLAPYVAIESVHDLLTGHR
MSMEG_5604|M.smegmatis_MC2_155 AGRD----------PEAREKVALRIVAFSFFGLATYVAVDAIRSLLGIGE
TH_1241|M.thermoresistible__bu AGRD----------PEARERIALRVIAFSFFGLATFVTVDAVRALLGMGE
MAB_2603c|M.abscessus_ATCC_199 SAKD----------PETREKVALRVIAFSFFALAAYVAVDSVRALAGLDQ
Mflv_3901|M.gilvum_PYR-GCK AGRD----------PEERERTALRIIAVSFFALATYVTVESARALFGAAE
MAV_3555|M.avium_104 RASRKLPTERYPYGYERAEDLAGIGVALVIWGSAVVAGFESVTKLLRHGG
. * : :*. :: * :: *
MMAR_1270|M.marinum_M VGTSVIGIGLTAFALVVMPILGRANHKLGVLLQSGATEGEGTQNYLCAGQ
MUL_2617|M.ulcerans_Agy99 VGTSVIGIGLTAFALVVMPILGRANHKLGVLLQSGATEGEGTQNYLCAGQ
Mb3299c|M.bovis_AF2122/97 AETSVIGIGLTAIALLLMPVLGWANHRVGERLGSGATAGEGTQNYLCAAQ
Rv3271c|M.tuberculosis_H37Rv AETSVIGIGLTAIALLLMPVLGWANHRVGERLGSGATAGEGTQNYLCAAQ
Mvan_3635|M.vanbaalenii_PYR-1 AEATVLGMVLAGSSLAVMPVLGYAKQRLGARLASGATAGEGIQNYLCAAQ
MSMEG_5604|M.smegmatis_MC2_155 AQHSTIGIALAALSLVIMPVLSYAQRRAGRELGSLSAVADSKQTLLCTYL
TH_1241|M.thermoresistible__bu SRPSTVGIVVAALSLLIMPVLSYAQRRAGRELGSLSAVADSKQTLLCTYL
MAB_2603c|M.abscessus_ATCC_199 ARQSTVGIALAAASLVIMPALSWAQRRTGRELGSASAVADSKQTLLCTYL
Mflv_3901|M.gilvum_PYR-GCK AAPSPAGIVLAAVSLAVMPFLSYAQRRAGRELGSASAVADSKQTLLCTYL
MAV_3555|M.avium_104 TGHVGWGIAAAVVGVVGNQLVARYKLVVGRRIRSATMVADAKHSWLDALS
*: : .: :. : * : * : .:. :. * :
MMAR_1270|M.marinum_M AAGVLLGLAITAAWPG----GWWIDPGIGLVIAGIAVWQGVRA-------
MUL_2617|M.ulcerans_Agy99 AAGVLLGLAITAAWPG----GWWIDPGIGLVIAGIAVWQGVRA-------
Mb3299c|M.bovis_AF2122/97 AAAVLLGLAITAVWSN----GWWIDPAIGLAIAGIAVWQGIRT-------
Rv3271c|M.tuberculosis_H37Rv AAAVLLGLAITAVWSN----GWWIDPAIGLAIAGIAVWQGIRT-------
Mvan_3635|M.vanbaalenii_PYR-1 AGAVLLSLAVTASWAGGWAGGWWLDPVIGLVIAAWSIWEGIQA-------
MSMEG_5604|M.smegmatis_MC2_155 SAVLLVGLLLNSIFGWS-----WADPIAGLVIAAVAVREGLEA-------
TH_1241|M.thermoresistible__bu SAVLLIGLLLNSLFGWS-----WADPVAGLAIAAMAVREGIEA-------
MAB_2603c|M.abscessus_ATCC_199 SAVLLAGLLVNSLFAWS-----WADPIAALAIAVIAVREGMNA-------
Mflv_3901|M.gilvum_PYR-GCK SGVLLVGLLLNALFGWS-----WADPIVALVIAGVAVKEGRQA-------
MAV_3555|M.avium_104 SAGAVLGLIG-VALGWG-----WADAVAGIVVTGFICHVGWEVTADIAHR
:. : .* * *. .:.:: * ..
MMAR_1270|M.marinum_M -------------------------------WRGQDCGC-----------
MUL_2617|M.ulcerans_Agy99 -------------------------------WRGQDCGC-----------
Mb3299c|M.bovis_AF2122/97 -------------------------------WRGHGCGC-----------
Rv3271c|M.tuberculosis_H37Rv -------------------------------WRGHGCGC-----------
Mvan_3635|M.vanbaalenii_PYR-1 -------------------------------WRGEDCCQ-----------
MSMEG_5604|M.smegmatis_MC2_155 -------------------------------WKGDTCCSAG---------
TH_1241|M.thermoresistible__bu -------------------------------WRGDTCCAVPVGT-SRTAT
MAB_2603c|M.abscessus_ATCC_199 -------------------------------WRGESCCMPVFAAGASELT
Mflv_3901|M.gilvum_PYR-GCK -------------------------------WRGEHCC------------
MAV_3555|M.avium_104 LLDGVDPDIVTTAEAVAVSVPGVTHAHARARWTGRTLRVEVEGFLDAATS
* *
MMAR_1270|M.marinum_M --------------------------------
MUL_2617|M.ulcerans_Agy99 --------------------------------
Mb3299c|M.bovis_AF2122/97 --------------------------------
Rv3271c|M.tuberculosis_H37Rv --------------------------------
Mvan_3635|M.vanbaalenii_PYR-1 --------------------------------
MSMEG_5604|M.smegmatis_MC2_155 --------------------------------
TH_1241|M.thermoresistible__bu ESGCGCGDDRE---------------------
MAB_2603c|M.abscessus_ATCC_199 GSQCGCCERRAP--------------------
Mflv_3901|M.gilvum_PYR-GCK --------------------------------
MAV_3555|M.avium_104 LSDSDRIGRSVAAALAPRLPQMQSFTWTARAA