For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLICAGSVVIDGELCRPGWIRTAGHKIADCGTGTAPLADFDFPDSIVVPGFVDIHVHGGAGASFADADAG GRALIQASEFHLRHGTTTMLASLVTAAPAQLLSAVAKLAEAISSGRSGTIAGIHLEGPWLSPARCGAHDH TQVRAPDPAEIDALLAAANGTIRMVTLAPELPGSADTIRRFLAADVVVALGHTDATYEQTCQAIGHGATV ATHLFNAMAPLGHREPGPALALLKDPAVTLELIADGVHVHPAVVAAVIEAVGPDRVALVTDAIAAAGCGD GTYRLGSVPIEVESNVARVCGTPTIAGSTATMDRLFRAAFRAGPGAALDAGALAAAVQMTSATPARAVGL ARRGSLRAGFDANLVVLDRDLRVAAVMANGEWQAVED*
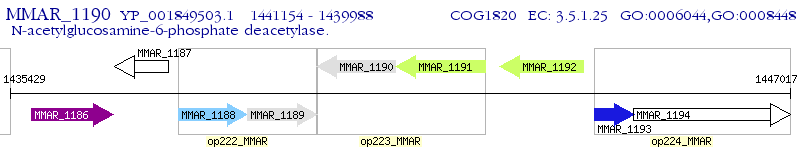
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1190 | nagA | - | 100% (388) | N-acetylglucosamine-6-phosphate deacetylase NagA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3365 | nagA | 1e-148 | 70.36% (388) | N-acetylglucosamine-6-phosphate deacetylase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3332 | nagA | 1e-148 | 70.36% (388) | N-acetylglucosamine-6-phosphate deacetylase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0362c | - | 9e-83 | 56.54% (306) | N-acetylglucosamine-6-phosphate deacetylase NagA |
| M. avium 104 | MAV_4307 | nagA | 1e-135 | 66.84% (386) | N-acetylglucosamine-6-phosphate deacetylase |
| M. smegmatis MC2 155 | MSMEG_2119 | nagA | 1e-100 | 52.36% (382) | N-acetylglucosamine-6-phosphate deacetylase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1443 | nagA | 0.0 | 98.97% (388) | n-acetylglucosamine-6-phosphate deacetylase NagA |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1190|M.marinum_M MTLICAGSVVIDGELCRPGWIRTAGHKIADCGTGTAPL-ADFDFPDSIVV
MUL_1443|M.ulcerans_Agy99 MTLICAGSVVIDGELCRPGWIRTAGHKIADCGTGTAPL-ADFDFPDSIVV
Mb3365|M.bovis_AF2122/97 MTVLGADAVVIDGRICRPGWVHTADGRILSGGAGAPPMPADAEFPDAIVV
Rv3332|M.tuberculosis_H37Rv MTVLGADAVVIDGRICRPGWVHTADGRILSGGAGAPPMPADAEFPDAIVV
MAV_4307|M.avium_104 MGLIAAGTMVASGRVHRPGWVETSGGHIAACGTGPPPRPADREFPDGTAV
MAB_0362c|M.abscessus_ATCC_199 ---------------------------------------------MSVTA
MSMEG_2119|M.smegmatis_MC2_155 -MLLTADTVLTGTELLRPGWLEIASDRVVAVGAGAPPAQADRNLGAATVV
. ..
MMAR_1190|M.marinum_M PGFVDIHVHGGAGASFADADAGGRALIQASEFHLRHGTTTMLASLVTAAP
MUL_1443|M.ulcerans_Agy99 PGFVDIHVHGGAGASFADADAGGRALIQASEFHLRHGTTTMLASLVTAAP
Mb3365|M.bovis_AF2122/97 PGFVDMHVHGGGGASFADGNAAD--IARAAEFHLRHGTTTTLASLVTAGP
Rv3332|M.tuberculosis_H37Rv PGFVDMHVHGGGGASFADGNAAD--IARAAEFHLRHGTTTTLASLVTAGP
MAV_4307|M.avium_104 PGFVDMHVHGGGGASYTD--PDG--IAKAAAFHLRHGTTTTMASLVTGSP
MAB_0362c|M.abscessus_ATCC_199 PGFFDMHVHGGGGASFGRDAEAN---LVAATWHRSHGTDGLLASLVTLAP
MSMEG_2119|M.smegmatis_MC2_155 PGFVDTHLHGGGGGNFSAATDDET--ARAVALHRAHGSTTLVASLVTAGP
***.* *:***.*..: * * **: :***** .*
MMAR_1190|M.marinum_M AQLLSAVAKLAEAISSGRSGTIAGIHLEGPWLSPARCGAHDHTQVRAPDP
MUL_1443|M.ulcerans_Agy99 AQLLSAVAKLAEAISSGRSGTIAGIHLEGPWLSPARCGAHDHTQVRAPDP
Mb3365|M.bovis_AF2122/97 AELLSAVGALAEAT---RDGVVAGIHLEGPWLSPARCGAHDHTRMRAPDP
Rv3332|M.tuberculosis_H37Rv AELLSAVGALAEAT---RDGVVAGIHLEGPWLSPARCGAHDHTRMRAPDP
MAV_4307|M.avium_104 AELLGGVRALAEAT---RDRTVAGIHLEGPWLSRARCGAHDARRMRDPDP
MAB_0362c|M.abscessus_ATCC_199 DDLLRAVRVLAEMT---GAEGIAGIHLEGPWLSPQYAGAHDPRLLREPDL
MSMEG_2119|M.smegmatis_MC2_155 EDLLRQVSGLARQV---RAGLIDGIHLEGPWLSTLRCGAHQPVLMRDPDP
:** * **. : ********** .***: :* **
MMAR_1190|M.marinum_M AEIDALLAAANGTIRMVTLAPELPGSADTIRRFLAADVVVALGHTDATYE
MUL_1443|M.ulcerans_Agy99 AEIDALLAAANGTIRMVTLAPELPGSADTIRRFLAAGVVVALGHSDATYE
Mb3365|M.bovis_AF2122/97 AEIESVLAAADGAVRMVTLAPELPGSDAAIRRFRDAEVVVAVGHTDATYT
Rv3332|M.tuberculosis_H37Rv AEIESVLAAADGAVRMVTLAPELPGSDAAIRRFRDAEVVVAVGHTDATYT
MAV_4307|M.avium_104 HEIDAVLSAGGGAIRMVTLAPELPGADAAIRRFVDAGVVVAVGHTNADYR
MAB_0362c|M.abscessus_ATCC_199 GELERLLDAGGGHIKMVTIAPELPGAISAIEMLSGRGVVAAVGHTNATYE
MSMEG_2119|M.smegmatis_MC2_155 GEIGRVLDAGEGTVRMVTIAPERDGALAAIAQLVNAGVVAAVGHTEATYD
*: :* *. * ::***:*** *: :* : **.*:**::* *
MMAR_1190|M.marinum_M QTCQAIGHGATVATHLFNAMAPLGHREPGPALALLKDPAVTLELIADGVH
MUL_1443|M.ulcerans_Agy99 QTCQAIGHGATVATHLFNAMAPLGHREPGPALALLKDPAVTLELIADGVH
Mb3365|M.bovis_AF2122/97 QTRHAIDLGATVGTHLFNAMPPLDHRAPGPVLALLCDPRVTVEIIADGVH
Rv3332|M.tuberculosis_H37Rv QTRHAIDLGATVGTHLFNAMPPLDHRAPGPVLALLCDPRVTVEIIADGVH
MAV_4307|M.avium_104 QTRHAIALGATVGTHLFNAMPPLHHREPGPALALLRDPRVTVELIADGVH
MAB_0362c|M.abscessus_ATCC_199 QTQQAISAGATVATHLFNAMRPIHHREPGPIPALLESPDVTIELIADGVH
MSMEG_2119|M.smegmatis_MC2_155 QTRAAIDAGATVGTHLFNAMRPIDRREPGPAVALTEDSRVTVEMIVDGVH
** ** ****.******* *: :* *** ** .. **:*:*.****
MMAR_1190|M.marinum_M VHPAVVAAVIEAVGPDRVALVTDAIAAAGCGDGTYRLGSVPIEVESNVAR
MUL_1443|M.ulcerans_Agy99 VHPAVVAAVIEAVGPDRVALVTDAIAAAGCGDGTYRLGSVPIEVESNVAR
Mb3365|M.bovis_AF2122/97 VHPAVVHAVIEAVGPDRVAVVTDAIAAAGCGDGAFRLGTMPIEVESSVAR
Rv3332|M.tuberculosis_H37Rv VHPAVVHAVIEAVGPDRVAVVTDAIAAAGCGDGAFRLGTMPIEVESSVAR
MAV_4307|M.avium_104 VHPDVVHAVIDAAGPERVALVTDALAAAGRPDGAFRLGPVHIDVVAGVAR
MAB_0362c|M.abscessus_ATCC_199 IHPAIYRTVLAAVGPDRIALVTDAMCAAGMPDGAYQLGDLPVTVAGGEAR
MSMEG_2119|M.smegmatis_MC2_155 VAPAIYRHITQTVGPERLSLITDAMAATGMSDGVYRLGPLDIDVVAGVAR
: * : : :.**:*::::***:.*:* **.::** : : * .. **
MMAR_1190|M.marinum_M VCGTPTIAGSTATMDRLFR-AAFRAGPG-AALDAGALAAAVQMTSATPAR
MUL_1443|M.ulcerans_Agy99 VCGTPTIAGSTATMDRLFR-AAFRAGAG-AALDAGALAAAVQMTSATPAR
Mb3365|M.bovis_AF2122/97 VAGASTLAGSTTTMDQLFR-TVAGLGSKSDSAGDVALAAAVQVTSATPAR
Rv3332|M.tuberculosis_H37Rv VAGASTLAGSTTTMDQLFR-TVAGLGSKSDSAGDVALAAAVQVTSATPAR
MAV_4307|M.avium_104 VHGTTTIAGSTATMDRLFR-TVAGSGTD----RDAALAAAVQMTSTTPAR
MAB_0362c|M.abscessus_ATCC_199 LP-NGTIAGSTASMADLHRFAAAQAGT----------DIATRQTSVNPRR
MSMEG_2119|M.smegmatis_MC2_155 VAGTDTIAGSTATMEQVFRLAVAHCGLP----RDDALSLAVRQACVNPAR
: *:****::* :.* :. * *.: :...* *
MMAR_1190|M.marinum_M AVGLARRGSLRAGFDANLVVLDRDLRVAAVMANGEWQAVED-----
MUL_1443|M.ulcerans_Agy99 AVGLARRGSLRAGFDANLVVLDRDLRVAAVMANGEWQAVED-----
Mb3365|M.bovis_AF2122/97 ALGLTGVGRLAAGYAANLVVLDRDLRVTAVMVNDDWRVG-------
Rv3332|M.tuberculosis_H37Rv ALGLTGVGRLAAGYAANLVVLDRDLRVTAVMVNDDWRVG-------
MAV_4307|M.avium_104 ALGLDRVGSLRAGHEANLVVLDDDWRVRAVMVRGDWLADP------
MAB_0362c|M.abscessus_ATCC_199 AVGC------------------------------------------
MSMEG_2119|M.smegmatis_MC2_155 ALGLPAAG-LAAGARADLVVLDHDLAVTAVMRAGEWVVTPGAAHTV
*:*