For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQAWDAICARRNVRQYLPEPVSEDDLNRIAEAGWRAPSAKNRQPWDFVIVTERSALQELSTVWMGAGHIA AAAAAIALVIPVPPDERRKVIDQYDVGQATMAMMIAATDLGIGTGHSSVGDQDKARAILGVPDDHLVAYL LGVGYPADRPLVPIRKPNRRAFAEVVHYGRW
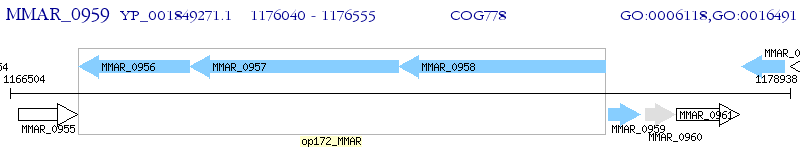
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0959 | - | - | 100% (171) | nitroreductase |
| M. marinum M | MMAR_0363 | - | 2e-10 | 29.61% (179) | hypothetical protein MMAR_0363 |
| M. marinum M | MMAR_4721 | - | 2e-08 | 29.06% (203) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0353 | - | 2e-09 | 27.36% (201) | cob(II)yrinic acid a,c-diamide reductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3351c | - | 4e-09 | 27.81% (151) | putative nitroreductase |
| M. avium 104 | MAV_4540 | - | 8e-82 | 82.46% (171) | nitroreductase family protein |
| M. smegmatis MC2 155 | MSMEG_2921 | - | 5e-08 | 27.56% (156) | NADPH-flavin oxidoreductase |
| M. thermoresistible (build 8) | TH_1618 | - | 5e-08 | 23.38% (201) | nitroreductase family protein |
| M. ulcerans Agy99 | MUL_0710 | - | 2e-69 | 99.21% (126) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_0381 | - | 4e-08 | 29.12% (182) | cob(II)yrinic acid a,c-diamide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0353|M.gilvum_PYR-GCK -----------MS-AETAALYDVINRRRDTRREFTGAPTDDEVLERVLLA
Mvan_0381|M.vanbaalenii_PYR-1 -----------MSDDSAAALYDVINRRRDTRREFTGEPVAGDVLQRVLLA
MAB_3351c|M.abscessus_ATCC_199 -----------------MDVYQAIATRRDVRAEFTGQVVDDATLMKLLQA
MMAR_0959|M.marinum_M -----------------MQAWDAICARRNVR-QYLPEPVSEDDLNRIAEA
MUL_0710|M.ulcerans_Agy99 --------------MGCMQAWDAICARRNVR-QYLPEPVSEDDLNRIAEA
MAV_4540|M.avium_104 -----------------MEAWDAICARRNVR-EYQPRAIAGEDLDRIVEA
TH_1618|M.thermoresistible__bu ------------MPHPTNDVWEVLSTARSIR-RFTDEPVDDATLQRCLQA
MSMEG_2921|M.smegmatis_MC2_155 MTVIARYADVDATLGVHSDTLALQLAHRSVR-KFLPDAVSDEHLSALVAA
*. * .: * *
Mflv_0353|M.gilvum_PYR-GCK AHAAPSVGMSQPWDFVLVRSPETLRAFRDHVAGEREVFAAELSGERADVF
Mvan_0381|M.vanbaalenii_PYR-1 AHAAPSVGMSQPWDFILVRSPQTLTTFRDHVAAERETFAGELSGERAETF
MAB_3351c|M.abscessus_ATCC_199 AHRAPSVGNSQPWDFVVIREPGMLDDFAAHVALQRQRFADSLPEGRRNTF
MMAR_0959|M.marinum_M GWRAPSAKNRQPWDFVIVTERSALQELSTVWMG-----AGHIAA------
MUL_0710|M.ulcerans_Agy99 GWRAPSAKNRQPWDFVLVTERSALQELSTVWMG-----AGHIAA------
MAV_4540|M.avium_104 GWRAPSAKNRQPWDFVIVTDRTQLQELSTVWRG-----AGHIAG------
TH_1618|M.thermoresistible__bu ATWAPNGANAQLWRFIVLDAPEQRAAVAEAAQIALHSIESIYGMSRPADD
MSMEG_2921|M.smegmatis_MC2_155 AQSAATSSNLQPWSVVAVRDPQRKARLAVLAKNQQFINDAPLFLVWVADL
. *.. * * .: : .
Mflv_0353|M.gilvum_PYR-GCK DRIKIEGICESGLGIVVGYDPTRGGPAVLGRHAIPDAGL--YSVVCAIQN
Mvan_0381|M.vanbaalenii_PYR-1 GRIKIEGICESGLGIVVGYDPTRAGPQVLGRHAVPDAGL--YSVVCAIQN
MAB_3351c|M.abscessus_ATCC_199 DPIKIEGIRESGTGVVVTYDHGRGGPHILGRHTISDAGL--FSAVLAIQN
MMAR_0959|M.marinum_M --------AAAAIALVIPVPPDE-------RRKVIDQ----YDVGQATMA
MUL_0710|M.ulcerans_Agy99 --------AAAAIALVIPVPPDE-------RRKVIDQ----YDVGQATMA
MAV_4540|M.avium_104 --------APAAIAIVVPEPPDE-------RRLVTDN----YDVGQATMA
TH_1618|M.thermoresistible__bu DRSRAARNNRATYELHDRAGEYTSVLFTAFKNEFSSELLQGGSIYPAMQN
MSMEG_2921|M.smegmatis_MC2_155 G-------RARRIAERAGVPLDG-------ADYLETTIIGFVDTALAAQN
. . *
Mflv_0353|M.gilvum_PYR-GCK LWLAATAEGLG---VGWVSFYREETLRSLVGMPDHVRPVAWLCV------
Mvan_0381|M.vanbaalenii_PYR-1 LWLAATVEGLG---VGWVSFYREEFLRSLVGMPDHVRPVAWLCV------
MAB_3351c|M.abscessus_ATCC_199 LWLAAVTEGIG---VGWVSFYEEQFLAELIDAPAHIRPIAWLCV------
MMAR_0959|M.marinum_M MMIAATDLGIG---TGHSSVGDQDKARAILGVPDDHLVAYLLGV------
MUL_0710|M.ulcerans_Agy99 MMIAATDLGIG---TGHSSVGDQDKAR-----------------------
MAV_4540|M.avium_104 MMIAATDLGIG---AGHSSVGDQDKARAILGVPDGHLVAFLLGV------
TH_1618|M.thermoresistible__bu FYLAARAQGLGACFTSWASYEGEKVLREAVGVPDDWFLAGHVVVGWPRGR
MSMEG_2921|M.smegmatis_MC2_155 AVLAAESLGLGTVFVGAIRNHPEEVAAELGLPPSAVATFGLAVGFPDPTE
:** *:* .. :.
Mflv_0353|M.gilvum_PYR-GCK --GPVADLPDVPDLERFGWQARRSLDTVVHRERYPEP-------------
Mvan_0381|M.vanbaalenii_PYR-1 --GPVAELPDVPDLERFGWRARSSLETVLHQECYRAR-------------
MAB_3351c|M.abscessus_ATCC_199 --GPVREFAERPDLERFGWRTGRPLEDAVHWGRYRA--------------
MMAR_0959|M.marinum_M --GYPADRPLVPIRK----PNRRAFAEVVHYGRW----------------
MUL_0710|M.ulcerans_Agy99 -------DPRCPRRP----PGR--LPAWRRISR-----------------
MAV_4540|M.avium_104 --GYPADRPLTPIRK----PNRRPFTEVVHRGRW----------------
TH_1618|M.thermoresistible__bu -HGPVRRRPLGDVVFRNRWDPDRADITYGRGARPKRR-------------
MSMEG_2921|M.smegmatis_MC2_155 NAGIKPRLPREAVLHHEQYDAQTADSHVPAYDERLADYNTRHGLTGTWSE
.
Mflv_0353|M.gilvum_PYR-GCK ------------------------------
Mvan_0381|M.vanbaalenii_PYR-1 ------------------------------
MAB_3351c|M.abscessus_ATCC_199 ------------------------------
MMAR_0959|M.marinum_M ------------------------------
MUL_0710|M.ulcerans_Agy99 ------------------------------
MAV_4540|M.avium_104 ------------------------------
TH_1618|M.thermoresistible__bu ------------------------------
MSMEG_2921|M.smegmatis_MC2_155 RVLARLAGPQSLSGRHLLRTQLERLGLGIR